Figure 1.
CAD/MI SNPs are enriched in regulatory regions of hepatocytes
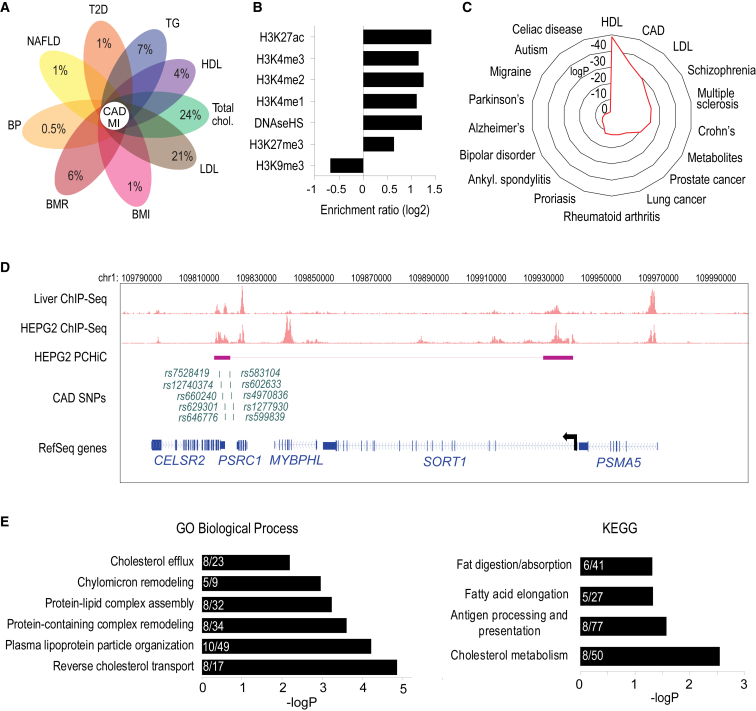
(A) Flower plot depicting the percentage of CAD/MI GWAS SNPs that are also associated with type 2 diabetes (T2D), triglycerides (TG), high-density lipoproteins (HDL), total cholesterol, low-density lipoprotein (LDL), body mass index (BMI), basal metabolic rate (BMR), blood pressure (BP), and nonalcoholic fatty liver disease (NAFLD) in the UK Biobank.
(B) Enrichment analysis of non-promoter regions in hepatocyte chromosomal interactions for enhancer- (H3K4me1-3, and H3K27ac) and repressor- (H3K27me3, H3K9me3) associated histone marks, and DNase I hypersensitive sites (DNaseHS).
(C) Radar chart showing the enrichment of GWAS variants within HepG2 chromatin interactions.
(D) Washu genome browser shot showing the location of SORT1 (chr1:109782257–109979272), H3K27ac ChIP-seq track for liver and HepG2, CAD-risk SNPs that fall within the looping ends, and PCHi-C interactions in HepG2 cells. Interacting restriction fragments are represented as boxes connected by a line on the HEPG2 PCHi-C track.
(E) Gene ontology analysis of the target genes identified from PCHi-C data.