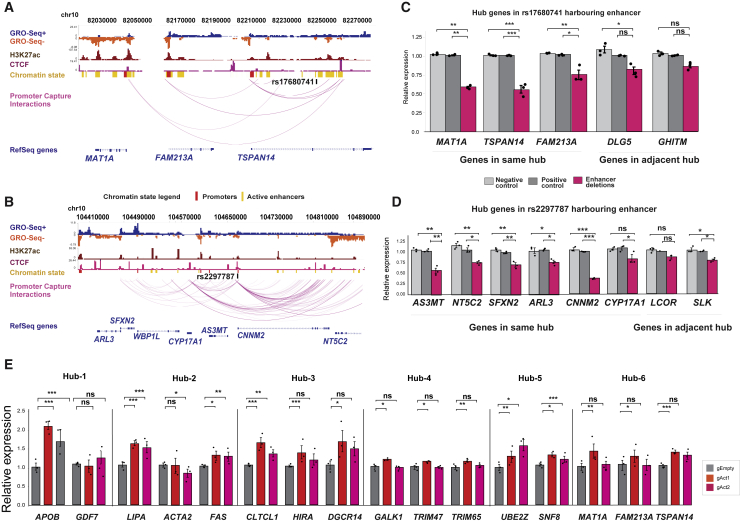
Figure 7.
CRISPR-mediated genetic perturbations of enhancer hubs
(A and B) Washu genome browser shots of two enhancer hubs containing (A) TSPAN14 and (B) SFXN2 where the CRISPR-Cas9 system was used to delete the enhancer region harboring a CAD GWAS variant. GRO-seq showing enhancer RNA (eRNA) for HEPG2 comes from GSE92375 (GSM2428726).
(C and D) Analysis of the effect of enhancer deletion on gene expression within the TSPAN14 and SFXN2 hubs in HepG2 cells. qPCR was performed for genes located in the same hub as well as for genes in adjacent hubs.
(E) Analysis of the effect of CRISPRa-mediated activation of enhancer variants in six selected chromatin hubs. For locus information, see Figure S9. Gene expression data are presented as the mean ± SEM of three independent experiments. The statistical significance was evaluated using a two-tailed Student’s t test or Mann-Whitney U test. For all bar plots, significance is denoted with asterisk. ∗p < 0.05, ∗∗p < 0.005, and ∗∗∗p < 0.0005.