Figure 7. Mapping of ELISpot positive peptides to the 500aa of the HBX2 HIV-1 gag protein sequence.
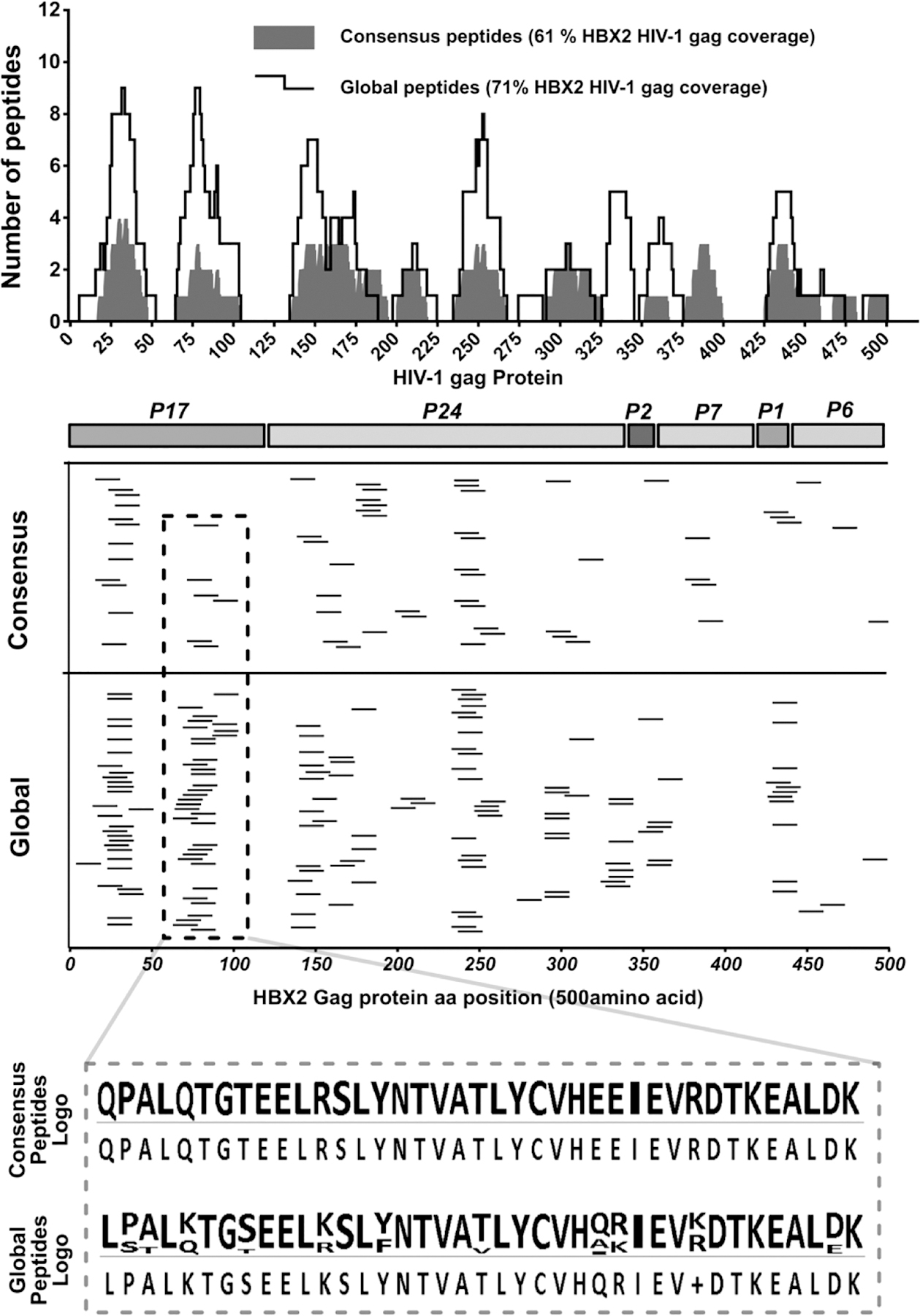
Epitopes were mapped in two subsequent ELISpot assays; the first ELISpot screened large peptide pools, while the second tested individual peptides. Determined response peptides were mapped to the HBX2 HIV-1 gag protein reference sequence to determine their relative locations. (Upper panel spectrum) Consensus and global peptides that elicited a positive ELISpot response with polyclonal expanded CD8 T cells were comparatively aligned to the HBX2 HIV-1 gag proteins. The number of peptides covering each gag protein sequence position was counted and plotted for each peptide set and the total coverage of the entire gag protein sequence determined. (Middle panels) Mapping of the positive ELISpot peptides from the consensus and global peptide sets to the HXB2 HIV-1 gag protein sequence positions and (lower panel) sequence logo of positive peptides from a select region on the Gag protein showing response global peptide sequence variations.
