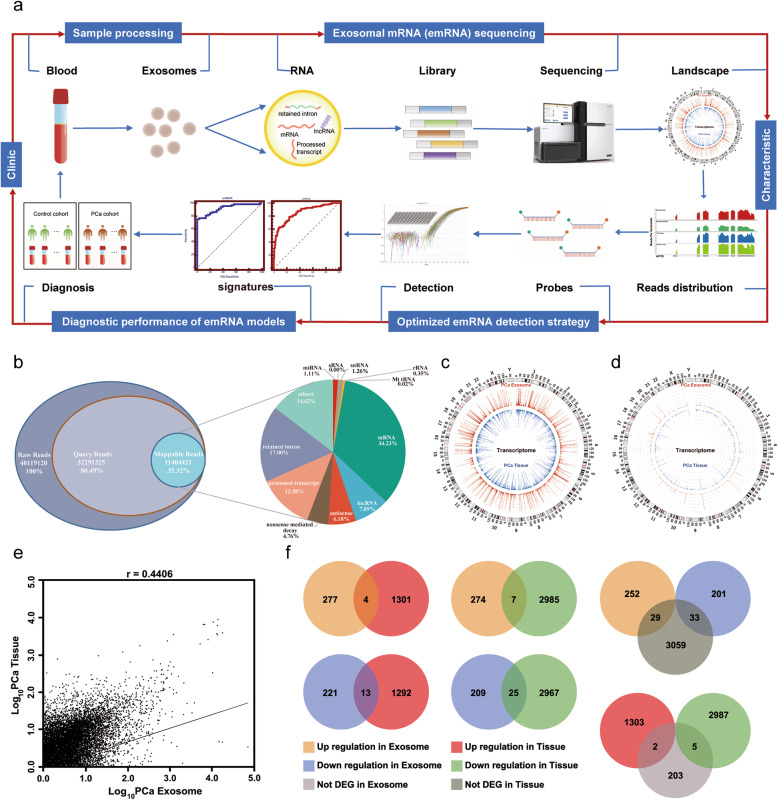
Fig. 1.
Characterization of circulating exosomal mRNAs (emRNAs). a, Workflow of the study, including sample processing, emRNA sequencing, demonstrating the landscape and characteristics of emRNA, optimizing the detection strategy, and identifying tumor-specific emRNA signatures. b, The type and distribution of RNAs in circulating exosomes. Raw reads are the sequences detected by RNA sequencing. Query reads are those after trimming. Mappable reads are those mapped to known human RNA or genomes. Circos plots showing all mRNAs (c), and oncogene mRNAs (d), from PCa tissues and circulating exosomes of the same cohort of patients. e, Scatter plot illustrating the correlation between tissue mRNA and emRNA levels. f, Venn diagrams showing the distinctive expression patterns between emRNAs and tissue mRNAs (based on the threshold of p value< 0.05 and fold change > 2 for upregulated and fold change < 0.5 for downregulated)