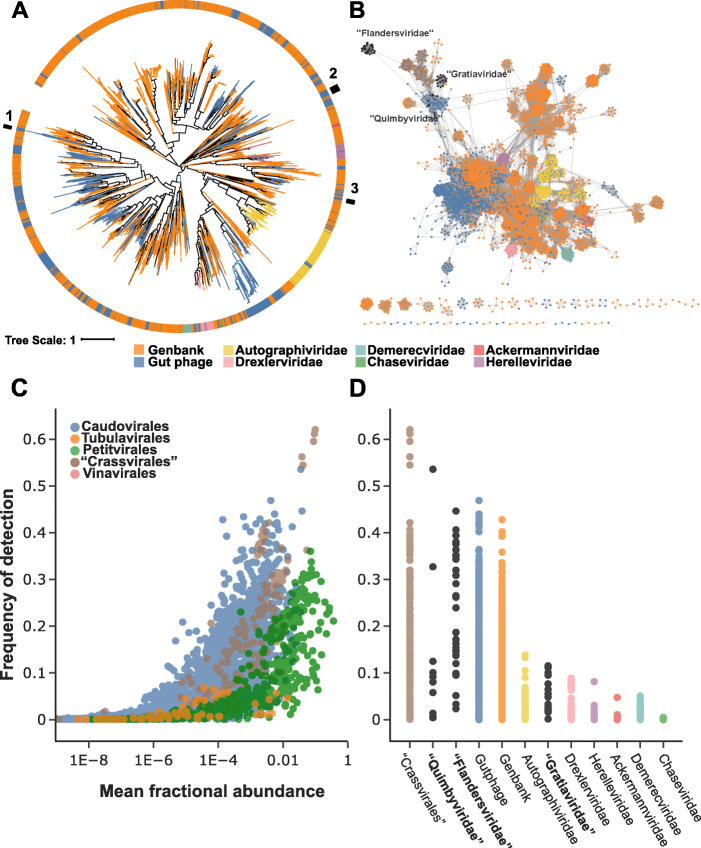
Fig. 1.
Three candidate families of Caudovirales phages discovered in human gut metagenomes. a Phylogenetic tree of the large terminase subunit encoded by Caudovirales phage genomes in GenBank (n = 3931) and in gut metagenomes (n = 1298). Branches are colored according to the current ICTV families, except for the Myoviridae, Podoviridae, or Siphoviridae, which are in orange. The outermost ring indicates the location of candidate families proposed in this study: 1, “Quimbyviridae” phages; 2, “Flandersviridae” phages; 3, “Gratiaviridae” phages (see main text). b Gene sharing network of the Urovicota phages. Phage genomes identified in human gut metagenomes (blue nodes) were compared to phages in the GenBank database (colored as in Figure 1, with the addition of the crAss-like phages in brown and the new Caudovirales families proposed in this study in black). c Abundance of phages across human gut viromes. The x-axis depicts the fractional abundance of a given phage averaged across all viromes (n = 1241); the y-axis is the fraction of viromes that a given phage recruits at least one read. Each phage genome (n = 7888 total) is colored at the taxonomic level of order (c) or family (Uroviricota families only) (d)