Fig. 1 |. RNA-targeting CRISPR-Cas (RCas) and parallel systems.
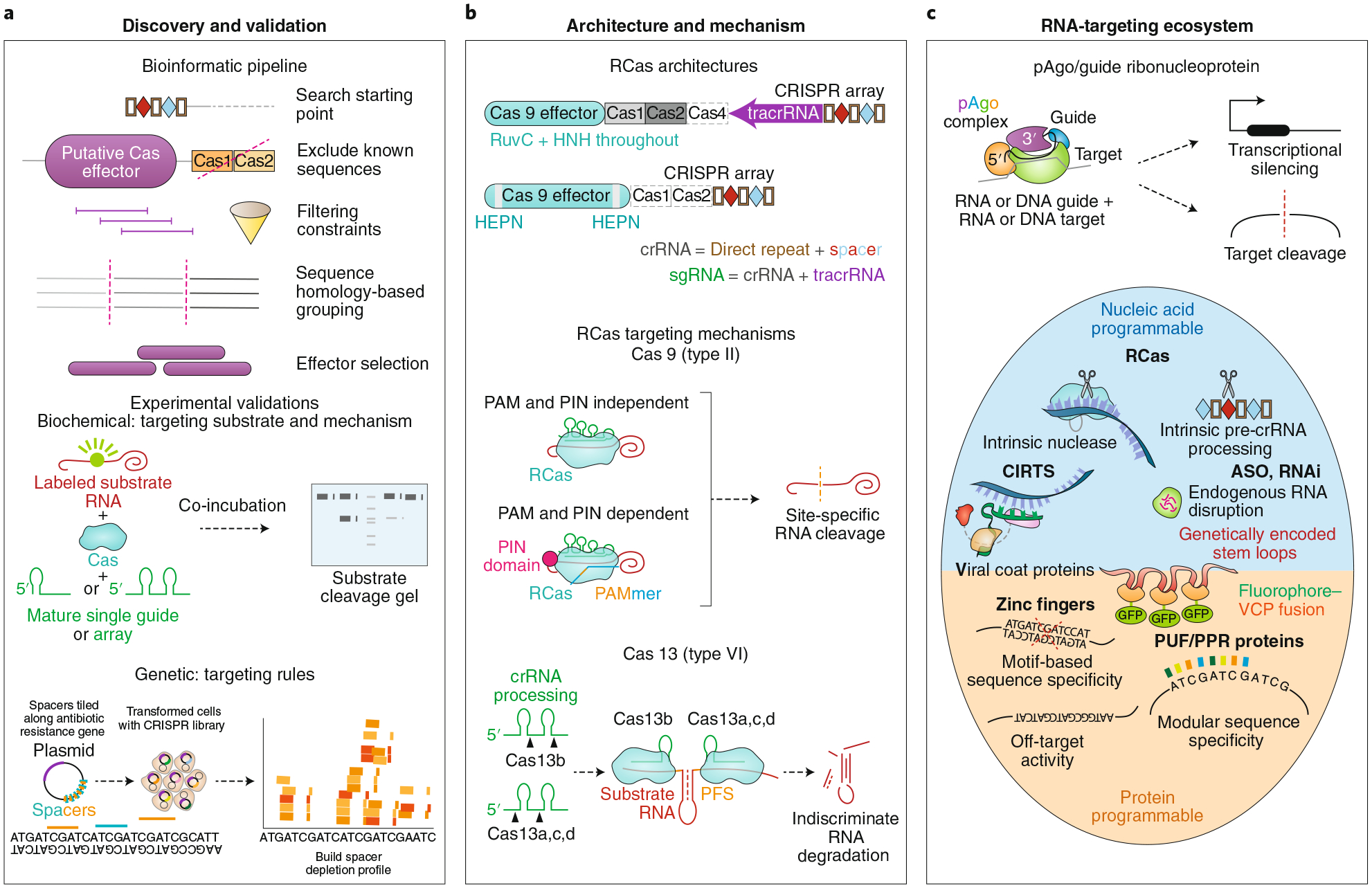
a, Cas proteins, including RCas, are discovered in metagenomic sequencing of bacteria and archaea using an established bioinformatic pipeline. Starting with a CRISPR array search seed, nearby putative Cas effectors are filtered on the basis of various criteria, including co-correlation with CRISPR arrays, size, and predicted protein domains. After grouping based on homology, distinct groups of effectors are selected for experimental validation follow-up. Researchers deduce the targeting substrate and mechanism through biochemical incubation of substrate nucleic acids with purified CRISPR ribonucleoprotein complexes and by observing resultant cleavage and binding. Genetic interrogation of targeting rules can be achieved through a spacer depletion screen assay consisting of a CRISPR library with spacers targeting an antibiotic resistance gene. Bacterial co-transformation via plasmids containing (i) the spacer library and Cas effector and (ii) the antibiotic resistance gene, followed by computational analysis of depleted spacers in the sequenced surviving population, will yield any targeting-dependent sequence requirements vis-à-vis the spacer. b, Type II and VI systems, encapsulating Cas9 and Cas13, respectively, represent the most commonly used RCas systems. Cas9 binds to a tracrRNA and crRNA (often combined into a single sgRNA) to target RNA. Depending on the Cas9 variant, a PAMmer oligonucleotide comprising a PAM sequence may be required for RNA binding10 and an additional domain (such as PIN domain endonuclease) may be required for RNA cleavage47. Cas13 binds to a crRNA for effective RNA targeting and subsequent indiscriminate cleavage. c, Prokaryotic Argonaute (pAgo) systems resemble CRISPR-Cas systems in their ability to target nucleic acids with a programmable guide. They exist as part of a broader RNA-targeting ecosystem that includes antisense oligonucleotides (ASOs), RNA interference (RNAi), CRISPR-Cas-inspired RNA targeting systems (CIRTS), viral coat proteins (VCPs), zinc fingers, Pumilio and FBF homology proteins (PUFs), and pentatricopeptide repeat proteins (PPRs), among others.
