Fig. 2 |. Applications of RCas in basic biology.
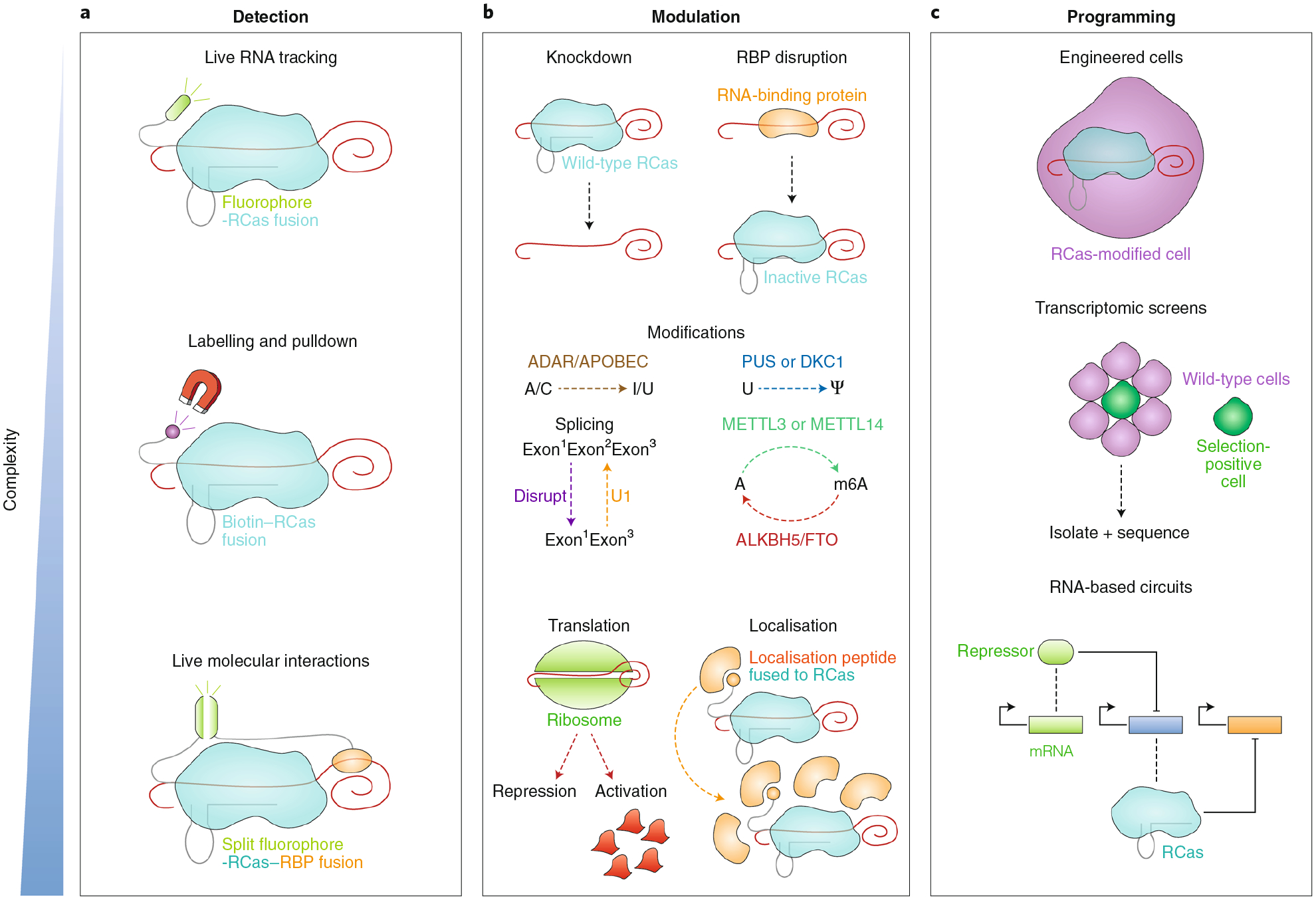
a, RCas can detect RNA in cells. Fusing a fluorophore to catalytically inactive RCas allows live-cell RNA tracking. Labelling the same RCas, for example, with biotin, followed by crosslinking enables pulldown of proximal proteins or RNA. Live protein-RNA interactions can be studied by fusing complementary split fluorophores to RCas and a protein. b, RCas can modulate RNA transcripts in cells. Expression of wild-type RCas results in RNA knockdown, whereas catalytically inactive RCas disrupts the binding of RBPs to RNA. Fusing an RNA-modifying effector to RCas enables the sequence modification of transcripts, such as A-to-I editing facilitated by the ADAR deaminase domain, U-to-pseudouridine editing by PUS (pseudouridine synthase) or DKC1 (dyskerin) enzymes, or m6A (methylation of adenine at the 6 position) by METTL3 or METTL14 enzymes. Other more challenging RNA modulations, such as translational activation/repression and localisation (by fusing a signal peptide to catalytically inactive RCas), could be achieved. c, RCas can be used to program cells. Researchers may engineer cells through any of the modulations previously described. RCas technologies can also be used to empower transcriptomic screens, facilitating selection of phenotypically positive cells among a pool of unperturbed wild-type cells. Finally, RCas can serve as a programmable element in RNA-based circuits, for example serving as a repressor for a transcript that in turn codes for an activating protein.
