Fig. 3 |. Opportunities for RCas in biotechnology.
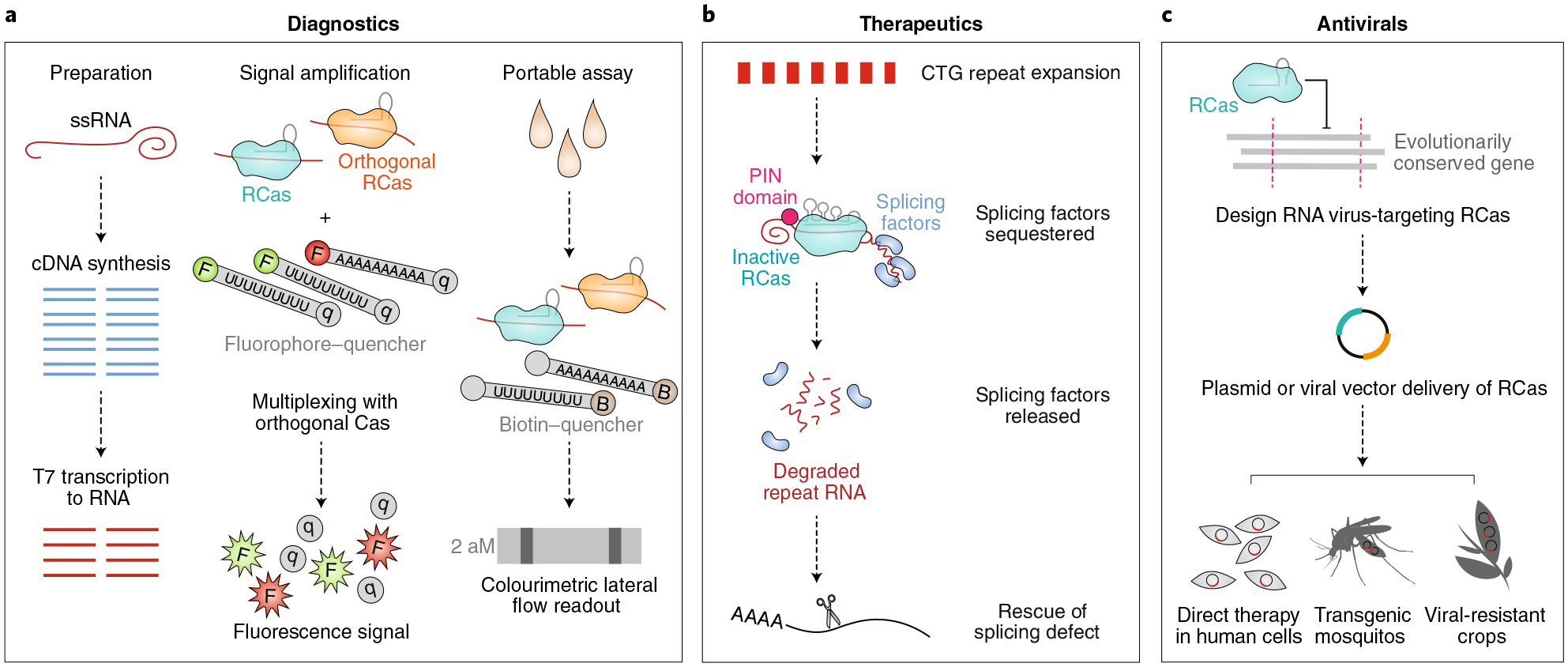
a, By leveraging the indiscriminate ssRNA cleavage of Cas13 upon target RNA recognition, researchers can quantitate the amount of any given nucleic acid species in a complex mixture. Briefly, RNA is converted into cDNA and then reverse transcribed back into RNA (or, alternatively, DNA is reverse transcribed into RNA). This amplified RNA is intermixed with short RNA fragments containing a fluorophore–quencher or biotin–quencher pair, as shown in the ‘signal amplification’ and ‘portable assay’ panels. Upon incubation of the mixture with Cas13 and a target-specific crRNA, fluorescence or colourimetric lateral flow readout corresponding exponentially to the concentration of a target RNA species can be quantitated for diagnostic purposes. b, RCas directed to CTG repeat expansions on RNA releases sequestered splicing factors by degrading the toxic repeat expansions, thus reversing associated RNA-splicing defects. Likewise, RCas can be applied to correct other RNA repeat expansions and RNA-related disorders in humans. c, By leveraging RCas to target evolutionarily conserved genomic regions of ssRNA viruses, researchers may invent antiviral human cell therapies, transgenic mosquitos with diminished capacity to spread infectious disease, and disease-resistant crops.
