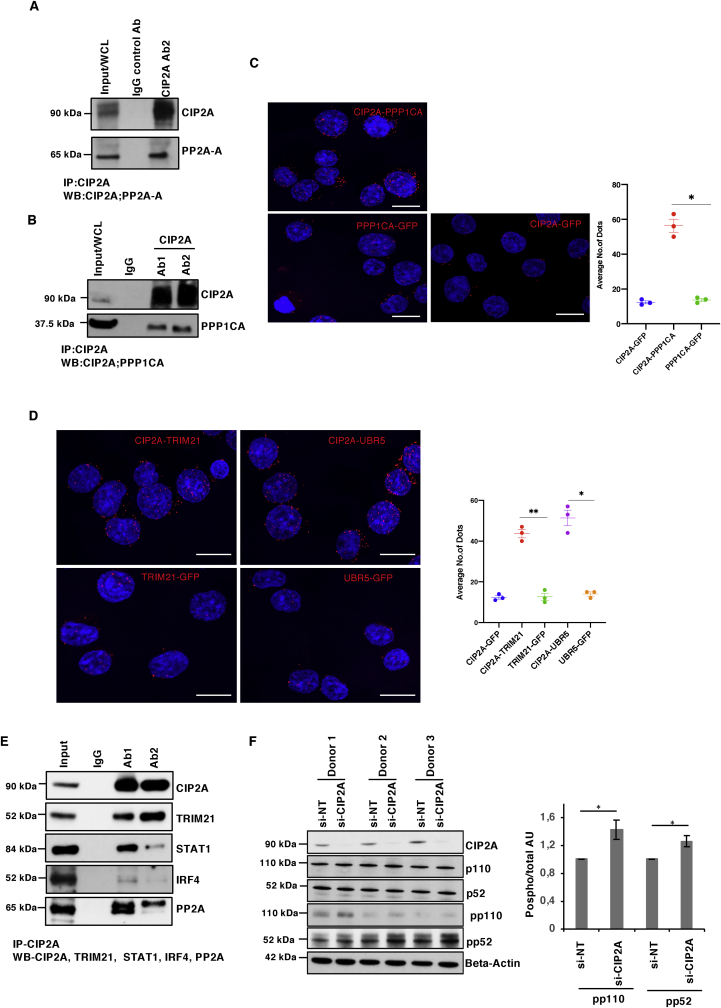
Fig. 5.
Validation of CIP2A protein interaction by IP-Western Blot and PLA Confocal microscopy. (A) CIP2A IP performed in Th17 cells and WB detection to validate the CIP2A known interaction with phosphatase PP2A-A. Total lysate (Input) and control IP (IgG) and CIP2A IP using Ab2 are shown on the blot. A conformational specific secondary antibody was used to probe proteins without interference from the denatured IgG heavy (50 kDa) and light chains (25 kDa). (B) WB validation of the CIP2A-PPP1CA interaction. CIP2A immunoprecipitated with two different antibodies (Ab1 and Ab2) in Th17 cells (72h) and detected as in A. (C-D) Proximity ligation assay (PLA) assay validation of the CIP2A interaction with PPP1CA (C) and TRIM21 and UBR5 (D) in Th17 cells (D). The GFP antibody was used as a negative control in the PLA. DAPI was used to stain the nuclei. Scale bar is 7μm. Representation of the interaction between CIP2A-PPP1CA (C) or TRIM21 and UBR5 with CIP2A (D) by PLA as a univariate dot plot. The average number of PLA signals (spots) per cell (n = 10 images from a representative of three experiments) was plotted. Statistics by paired two tailed T-test, where ∗ represents P < 0.05. and ∗∗< 0.01. Scale bar is 7μm. (E) Western blot validations representative of interactions between TRIM21, IRF4, STAT1 and PP2A with CIP2A in Th17 cells at 72h. IP was performed using Ab1 and Ab2 CIP2A antibodies and a conformational specific secondary antibody was used to detect the protein as in Fig. 5A. A representative Western blot of four biological replicates is shown, the remaining three Western blot replicates are shown in Figs. S5B–D. (F) CIP2A inhibition leads to enhanced Nf-κB activation. CIP2A is silenced in three different individual donor cell pools, and CIP2A, phospho (pp52 and pp110) and total (p52 and pp110) Nf-κB2 and Actin are shown on the blot. The bar chart on the right shows the quantification of the phosphorylation levels of p110 and p52 in CIP2A silenced Th17 cells. Quantification was performed using ImageJ software in which phosphorylation values were normalised with respective total protein p110 and p52 expression. Statistical significance calculated using Student's t-test (two-tailed paired) where ∗ denotes p-value <0.05.