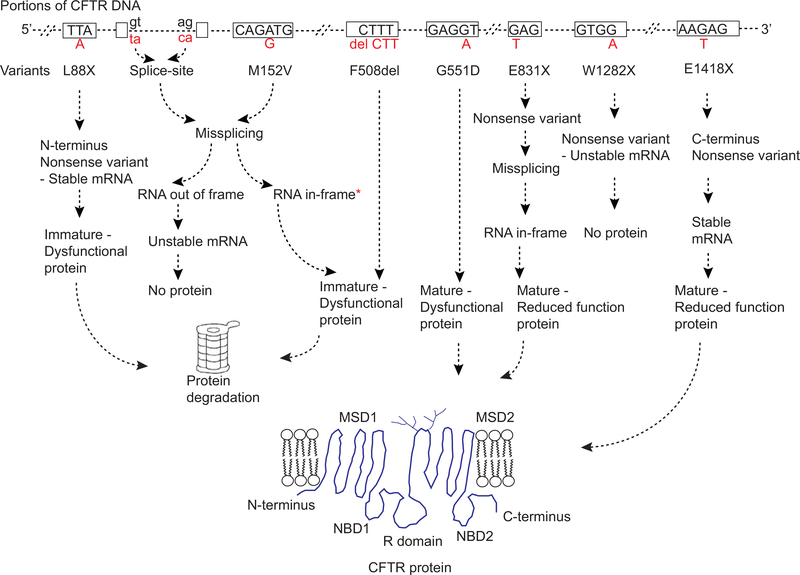
Fig. 1.
Genetic variants in CFTR and their impact on RNA and protein production. Only a portion of CFTR gene is shown. Rectangular boxes are exons, dashed lines are either introns, 5′UTR or 3′UTR, and dashed hashes are multiple exon-introns within CFTR gene. Please note, only few nucleotides within each exon are shown. Dinucleotides, gt and ag, in black letters represent consensus splice site signals. Alphabets in red beneath each exon or intron represent nucleotide changes. Variant names are written beneath each nucleotide change. Nonsense and frameshift variants can have heterogeneous effects on mRNA stability depending upon their location [13]. Misfolded CFTR protein undergoes ER-associated degradation via the ubiquitin–proteasome system. Fully glycosylated mature protein can be dysfunctional due to impaired gating, conductance or reduced residence time at the cell surface [4]. Missense variants predicted to make mature protein can undergo mis-splicing resulting in no protein or immature dysfunctional CFTR protein [9–11]. *indicates in-frame mis-spliced mRNA may also generate mature but dysfunctional CFTR protein.