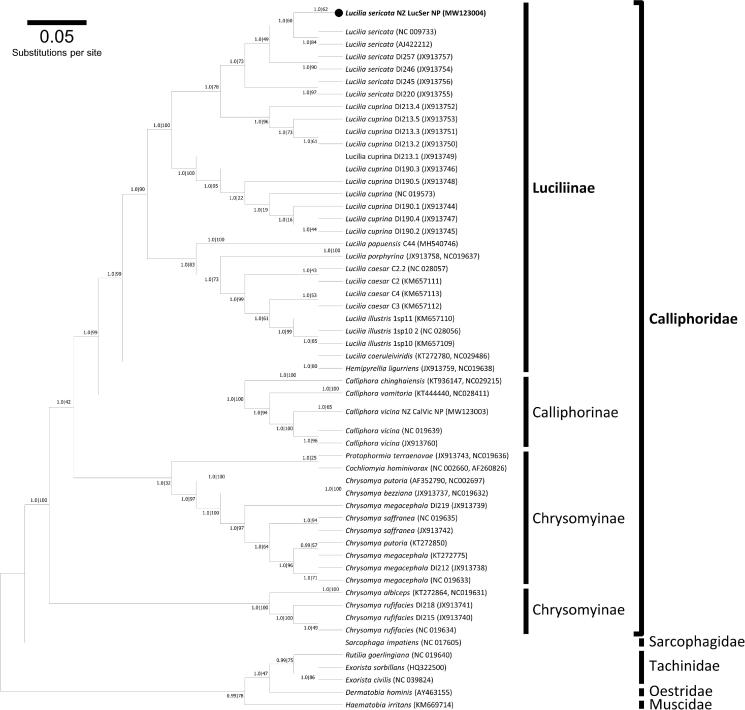
Figure 1.
A summary of the molecular phylogeny of the Calliphoridae complete mitochondrial genomes. The evolutionary relationship of Lucilia sericata field strain NZ_LucSer_NP (black circle) was compared to the complete mitochondrial genomes of 67 blowfly species or isolates retrieved from GenBank (accession numbers in parentheses) and nucleotide sequences of all protein-coding genes were used for analysis. Phylogenetic analysis was conducted using the Bayesian approach implemented in MrBayes version 3.2.6 (Huelsenbeck and Ronquist 2001) and maximum likelihood (ML) using RAxML version 8.2.11 (Stamatakis 2014). The mtREV with Freqs. (+F) model was used for amino acid substitution and four independent runs were performed for 10 million generations and sampled every 1000 generations. For reconstruction, the first 25% of the sample was discarded as burnin and visualized using Geneious Prime (Kearse et al. 2012). Nodal support is given: Bayes posterior probabilities RAxML bootstrap percentage. The phylogram provided is presented to scale (scale bar = 0.05 estimated number of substitutions per site) with the species Haematobia irritans from the family Muscidae used as the outgroup.