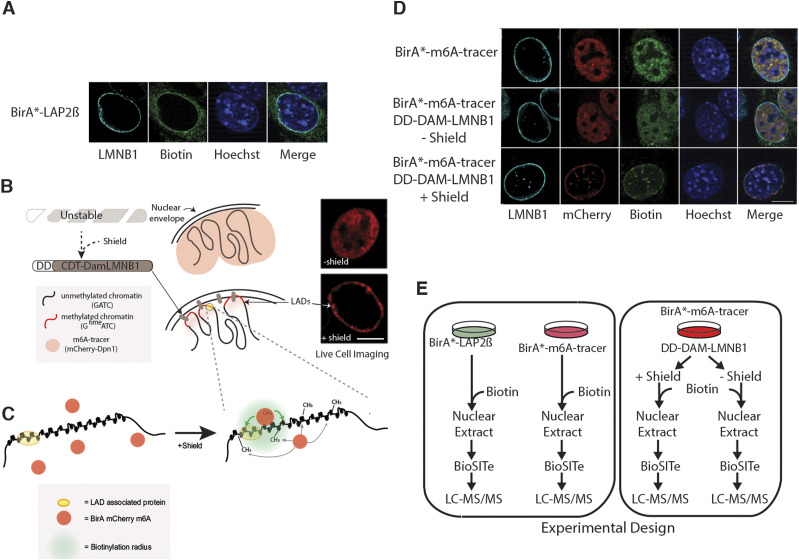
Figure 1. Strategy for investigating the nuclear periphery microproteomes.
(A) Representative images showing the biotinylated interactome of Lap2β (green) at the nuclear periphery in mouse embryonic fiboroblasts (MEFs). (B, C) Pictorial representation of BirA* localization within the nucleus. Lamina-associated domains become methylated at GATCs upon Shield1 stabilization of Dam LmnB1, thereby recruiting BirA* mCherry m6A. Green shading in (C) depicts a putative biotinylation radius where available lysines on proximal proteins will be modified by BirA*. (D) Representative images showing the expected localization of the BirA*-m6a-tracer (red) and its biotinylated proteins (green) in the absence of DD-Dam LmnB1 (top row), in the absence (middle row) and presence (bottom row) of the shield ligand in MEFs. (E) Experimental workflow for mass spectrometry based BioID lamina associated domain microproteome analysis. MEF cells expressing BirA*-LAP2β, BirA*-m6A-tracer alone, BirA*-m6A-tracer/DD-Dam-LMNB1 plus/minus shield ligand were cultured with exogenous biotin then nuclei were extracted and Biotinylation Site Identification Technology (BioSITe) coupled to liquid chromatography/tandem mass spectrometry analysis was used to identify biotinylated proteins.