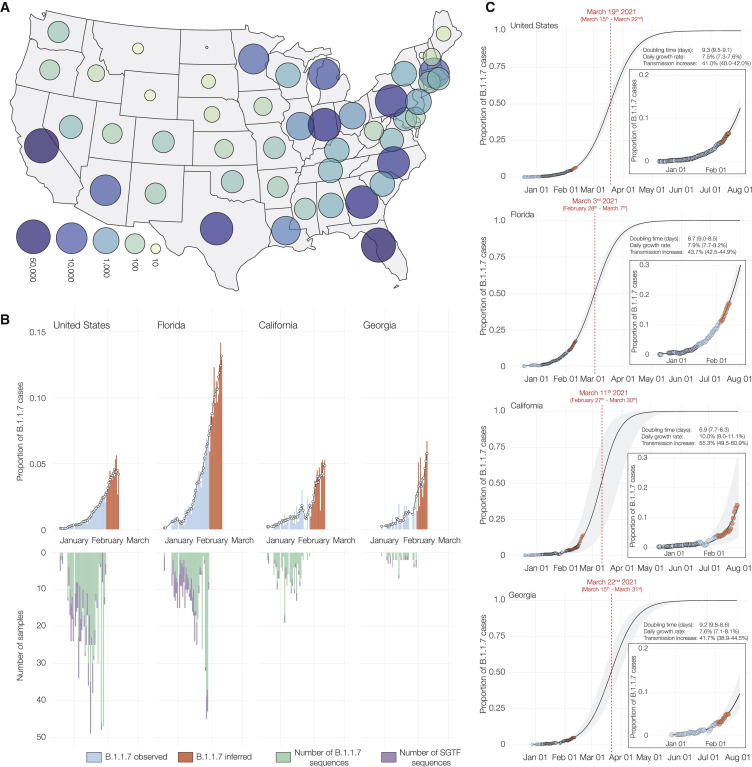
Figure 1.
Estimated proportion of B.1.1.7 in SARS-CoV-2 tests at Helix since December 15, 2020
(A) Map of contiguous states in the US with each bubble representing the number of positive Helix COVID-19 tests from each state.
(B) Estimated proportion of B.1.1.7 in total number of positive tests with Cq(N gene) <27, in the US overall, California, Florida, and Georgia from December 15th, 2020 to February 11th, 2021. The proportion of B.1.1.7 samples was estimated using: . There is an ∼2 week lag between sequence data and testing data. We had sequence data until February 2nd, but we had testing data until February 19th. To fully utilize the testing data, we used the average proportion of B.1.1.7 sequences in sequenced samples with SGTF from the last 5 days of available sequence data in each location to infer the proportion of B.1.1.7 cases in total positive tests for an additional 2 week period (February 3 to February 19). The black line shows the 5-day rolling average of the estimated proportion of B.1.1.7 in total positives. The inverted bar chart shows the temporal distribution of the B.1.1.7 genomes sequenced and the number of sequenced samples with SGTF.
(C) Logistic growth curves fit to the rolling average of the estimated proportion of B.1.1.7 in total positives for the US, Florida, California, and Georgia. The shaded area represents the 95% CI for each fit. The inset shows the zoomed in view of the curve fit. The predicted time when the estimated proportion of B.1.1.7 cases crosses 0.5 is indicated in red. See also Data S1.