Fig. 6. ChIP-seq analysis.
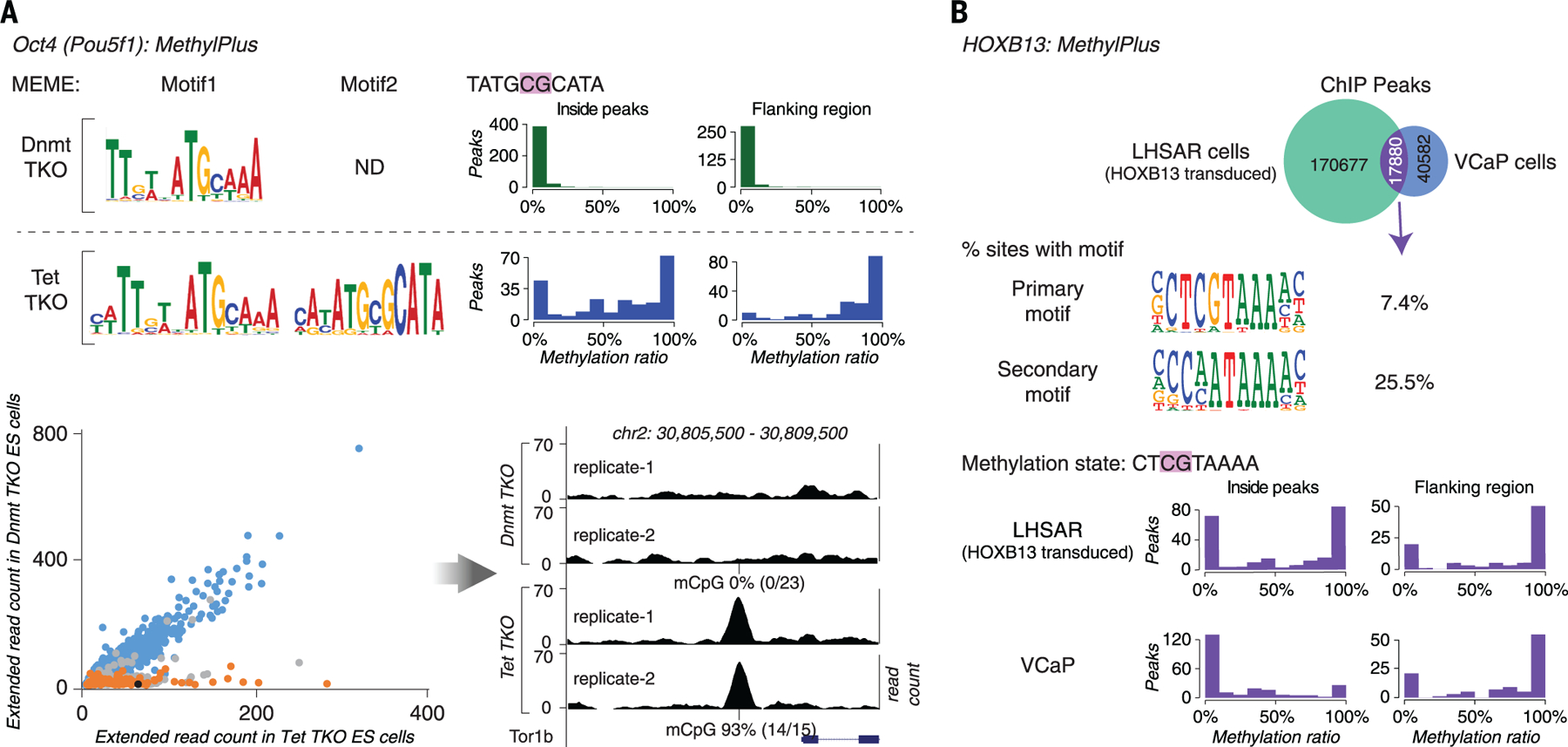
(A) OCT4 prefers a methylated motif in vivo. ChIP-seq analysis of OCT4 was performed in ES cells lacking methylcytosine (Dnmt-TKO) or displaying increased methylation of gene regulatory regions (Tet-TKO). Motif enrichment analysis with MEME (top left) recovered the methyl-plus motif (motif 2) of OCT4 only from peaks from the Tet-TKO cells. Most of the OCT4 occupied sites containing motif 2 were fully methylated in Tet-TKO cells (blue histograms) but not in Dnmt-TKO cells (green histograms) (top right). The scatterplot (bottom left) shows ChIP extended read coverage at motif match positions from peaks in the Tet- (x axis) and Dnmt-TKO (y axis) cells. The ChIP-seq peak heights at the motif 1 match positions (blue) are similar among the cell types, whereas peaks at motif 2 match positions whose methylation state changes (orange) are taller in Tet-TKO cells. Only sites that overlapped with two or more bisulfite-sequencing reads were analyzed. Peaks containing motif 2 matches whose methylation does not change or changes less than the cut-off (from ≤ 20% in Dnmt-TKO to ≥80% in Tet-TKO) are in gray. The black dot indicates the example peak site shown in the bottom right panel. (B) Exogenously introduced HOXB13 binds to methylated sites in the primary prostate epithelial cell line LHSAR. ChIP-seq analysis for HOXB13 was performed in VCaP prostate cancer cells and LHSAR cells transduced with HOXB13-expressing lentivirus. Analysis of peaks that were common to both cell lines (top) showed that HOXB13 can bind to two different motifs, one of which (SELEX primary motif) commonly contains a CpG dinucleotide. The positions of most of the common peaks containing CpG were methylated in LHSAR cells, indicating that HOXB13 can bind to methylated sites. The methylation level of the occupied sites is generally either very low or very high, consistent with the fact that methylation is either present or absent at a given allele. Methylation is lower in the VCaP prostate cancer cells, potentially because of binding-induced demethylation (7).
