Extended Data Fig. 7. Workflow for preparation of standard samples to assess quantitative DI-SPA.
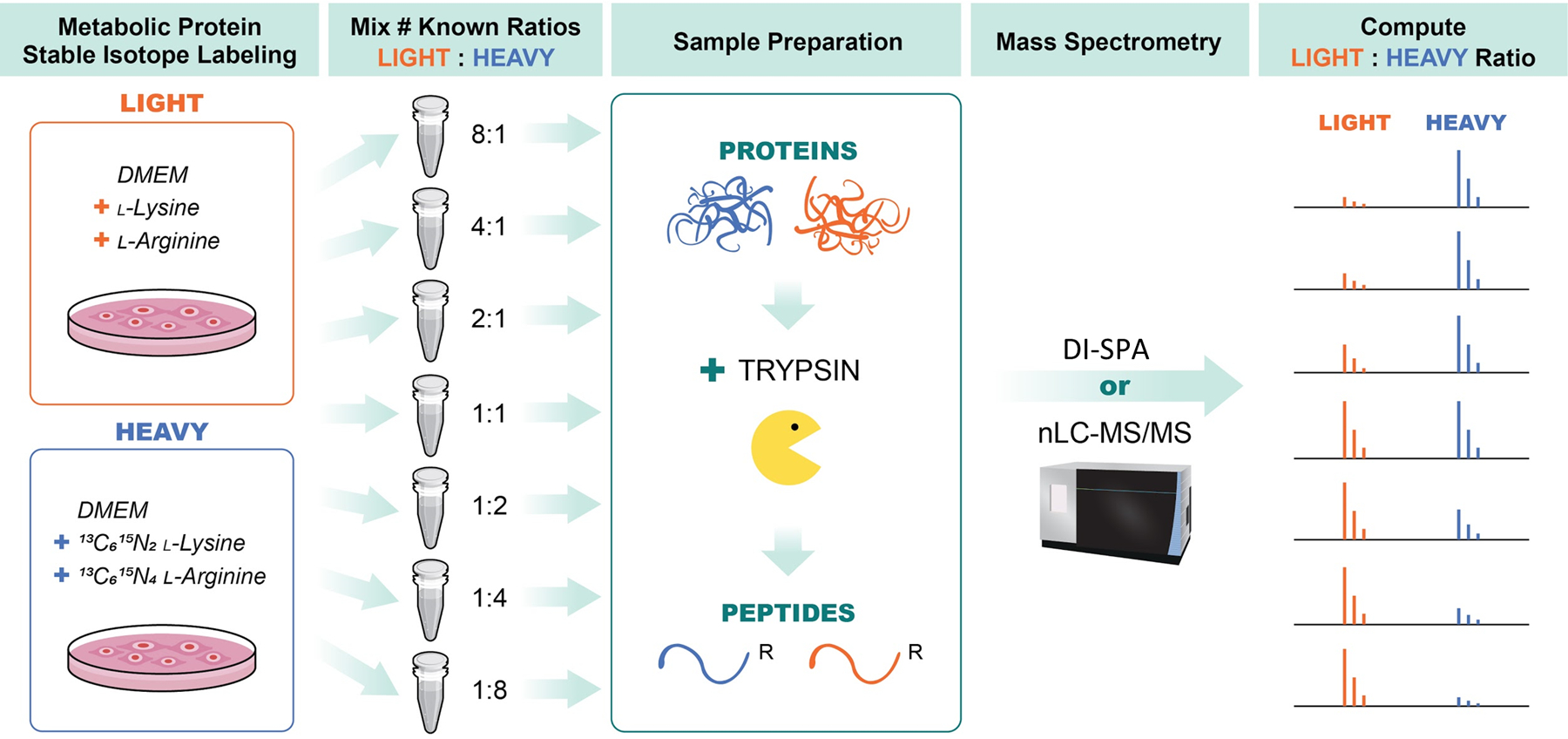
A549 cells were grown in DMEM media containing either light lysine and arginine (LIGHT) or 13C6, 15N2 -lysine and 13C6,15N4 L-arginine (HEAVY) and then combined at various ratios including: 1:8, 1:4, 1:2, 1:1, 2:1, 4:1, and 8:1 (HEAVY:LIGHT). Samples were then lysed proteins were reduced and alkylated, and proteolysis was initiated with trypsin. Peptides from trypsin digestion were desalted and then data was collected in parallel with either traditional nanoLC-MS/MS to verify SILAC ratios and provide a benchmark, or with DI-SPA to determine quantitative quality. Data from nanoLC-MS/MS was analyzed using MaxQuant to identify and quantify peptides, and data from DI-SPA was analyzed with MSPLIT-DIA and custom code in python and R.
