Extended Data Fig. 8. Examples of relationships between DI-SPA data collection settings for different peptides and their corresponding proteins.
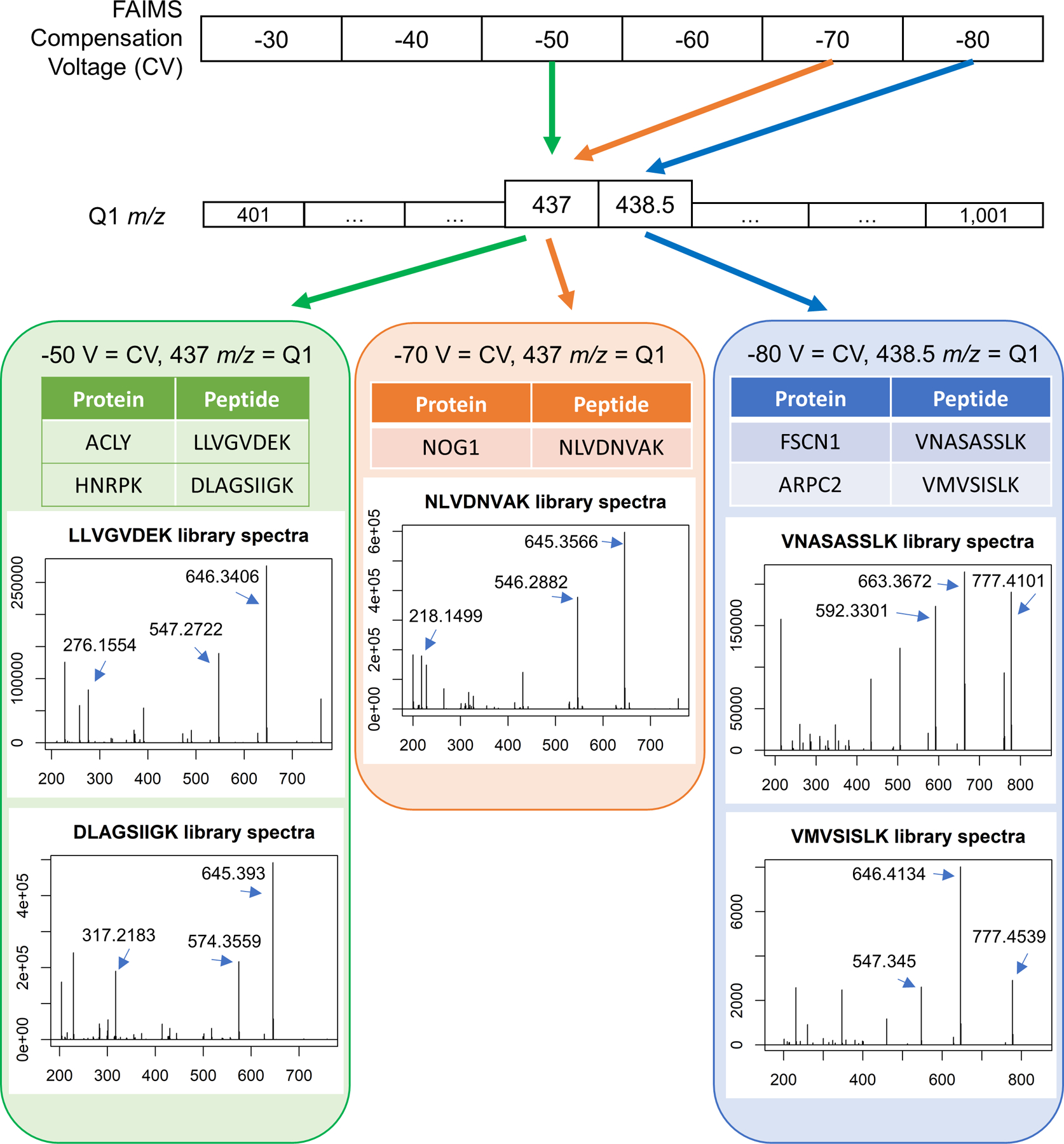
Peptides that uniquely identify proteins are found with a combination of gas-phase fractionation by FAIMS and precursor mass isolation with the first quadrupole (Q1). In this example, two unique peptides from different proteins are co-isolated with FAIMS compensation voltage (CV) of −50 V and Q1 set to 437 m/z. A single peptide is isolated with CV of −70 V and the same Q1 setting of 437 m/z, and two more peptides are isolated with a CV of −80 V at Q1 set to 438.5 m/z. Library spectra are shown for each peptide. The three most abundant singly charged y-ions in the library spectra are used for peptide quantification unless otherwise noted.
