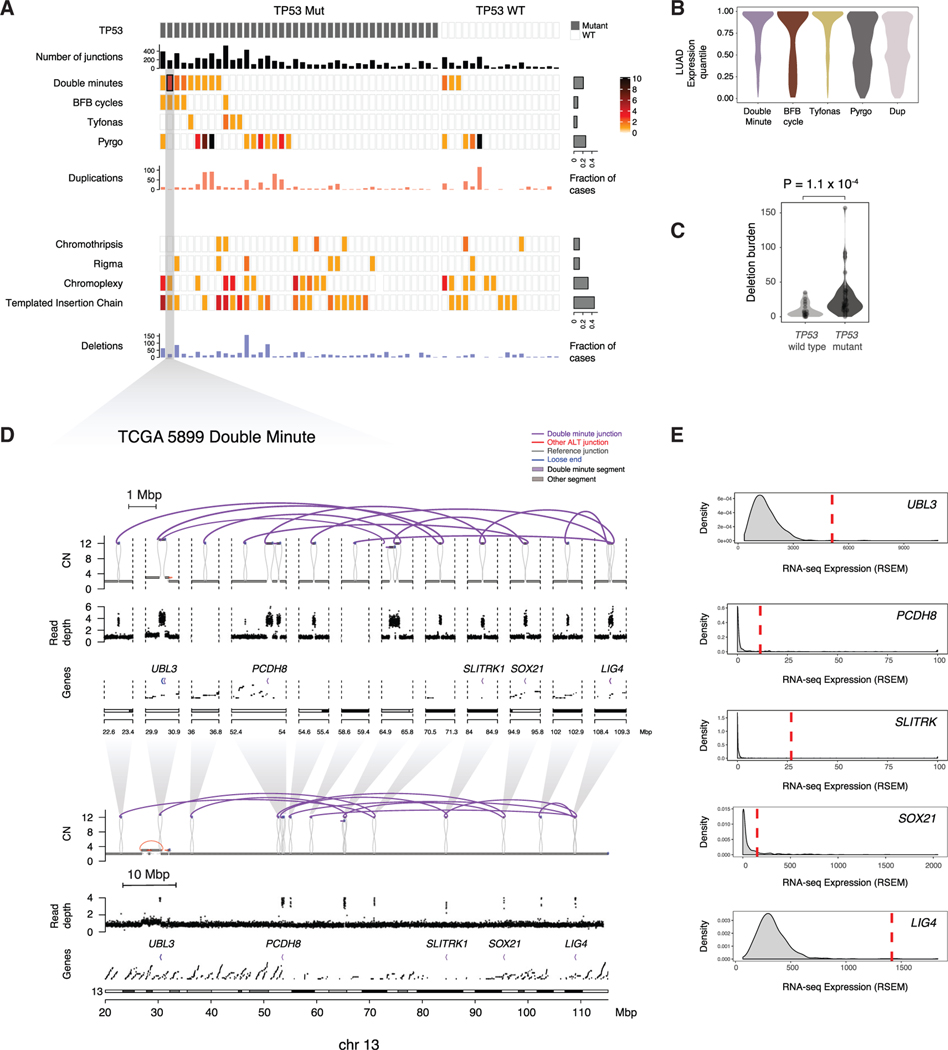
Figure 4. Classification of SVs in RPA(–)G LUADs.
(A) Identification of simple and complex SV events. Upper panel: SVs resulting in copy-number gain (double minute, BFBC, tyfonas, pyrgo, simple duplication). Lower panel: SVs resulting in copy-number loss (chromothripsis, rigma, chromoplexy, templated insertion chain, simple deletion). Key indicates the range of event count of SV types observed in each sample.
(B) Expression quantile of genes located in SV types with copy-number gain.
(C) Simple deletion count is more significantly enriched in the TP53 mutant RPA(–)G samples than in the TP53-wild-type RPA(–)G samples. p value is obtained from Mann-Whitney U test. Violin plots reflect kernel density estimations.
(D and E) Example of a double minute in TCGA-55–5899 spanning 3 genes (D), and (E) 2 of which (UBL3 and LIG4) showed marked overexpression relative to RNA-seq data for the full TCGA LUAD cohort. See also Figure S4.