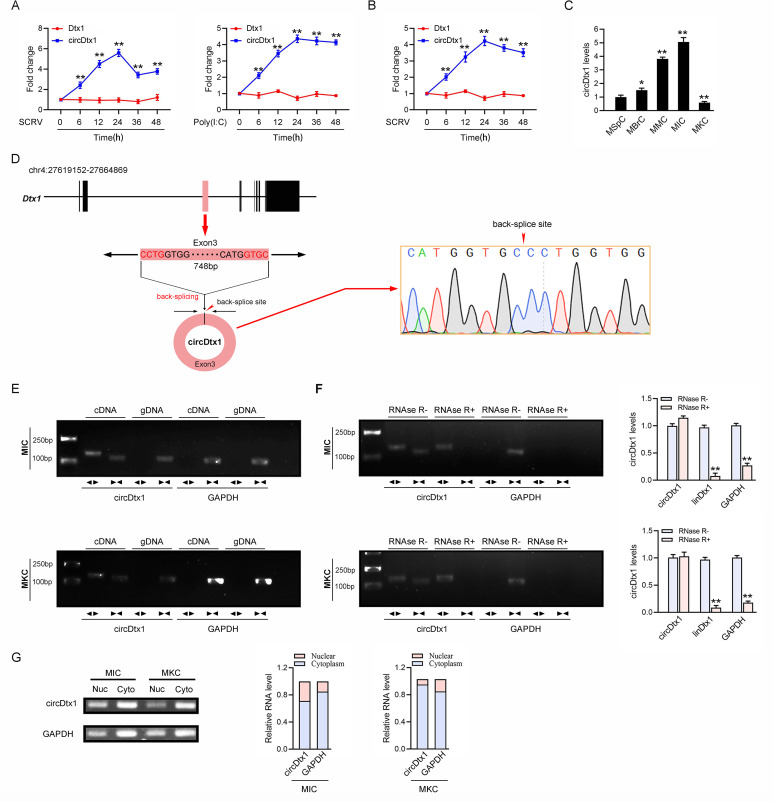
Fig 1. Expression profiles and characterization of circDtx1.
(A) qPCR for the abundance of circDtx1 and linear Dtx1 (Dtx1) mRNA in spleen tissues treated with SCRV (MOI = 5) and poly(I:C) at the indicated time points, respectively. (B) qPCR analysis of circDtx1 and linear Dtx1 mRNA in MKC cells treated with SCRV (MOI = 5) at the indicated time points. (C) Relative expression of circDtx1 in indicated cell lines was determined by qPCR. (D) We confirmed the head-to-tail splicing of circDtx1 in the circDtx1 RT-PCR product by Sanger sequencing. (E) RT-PCR validated the existence of circDtx1 in MIC and MKC cell lines. circDtx1 was amplified by divergent primers in cDNA but not gDNA. GAPDH was used as a negative control. (F) The expression of circDtx1 and linear Dtx1 mRNA in both MIC and MKC cell lines was detected by RT-PCR assay followed by nucleic acid electrophoresis or qPCR assay in the presence or absence of RNase R. (G) circDtx1 was mainly localized in the cytoplasm. RNA isolated from nuclear and cytoplasm was used to analyze the expression of circDtx1 by RT-PCR; n = 3. The data represented the mean ± SD from three independent triplicated experiments. **, p < 0.01.