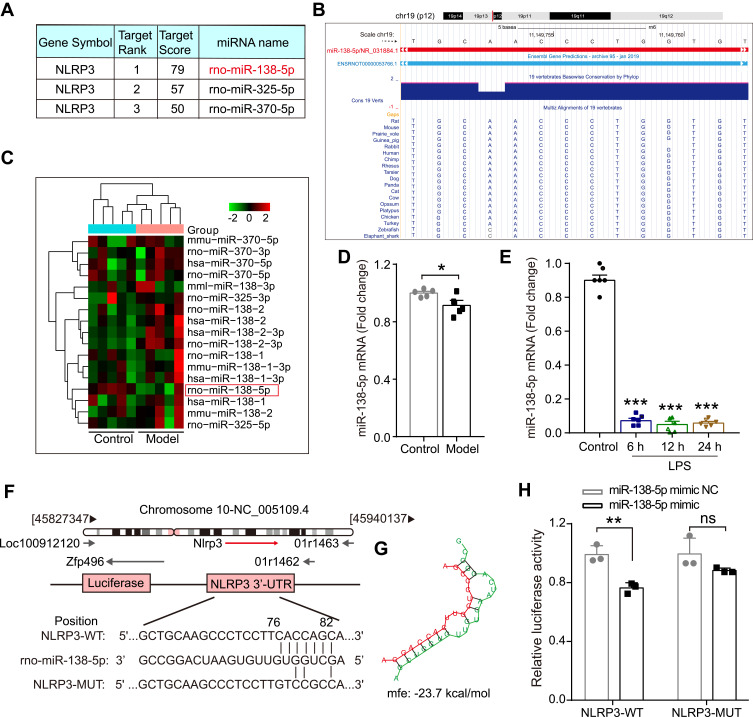
Figure 3.
The expression of miR-138-5p in the hippocampus of LPS-treated rats. (A) Three miRNAs that could bind to NLRP3 in rats predicted by miRDB software. (B) The basic information of chromosome location, base sequence and species conservativeness of miR-138-5p obtained from UCSC Genome Bioinformatics website. (C) Heatmap for the three predicted miRNAs expression and (D) relative miR-138-5p level in the hippocampal of the cognitive impairment animal model and the control group (n = 5 pre group) from NCBI GEO database. (E) The qRT-PCR result of miR-138-5p mRNA expression in the LPS-treated rat hippocampus (n = 6 pre group). (F) The predicted binding sites and (G) mfe between miR-138-5p and the 3ʹ-UTR of NLRP3. (H) A firefly luciferase reporter containing NLRP3-WT/MUT with miR-138-5p mimic NC/mimic (n = 3 pre group). Data were presented as mean ± SEM. *p < 0.05, **p < 0.01.
Abbreviations: GEO, Gene Expression Omnibus; LPS, lipopolysaccharide; NCBI, National Center for Biotechnology Information; GEO, Gene Expression Omnibus; mfe, minimum free energy; WT, wild-type; MUT, mutant; NC, negative control; ns, not significant.