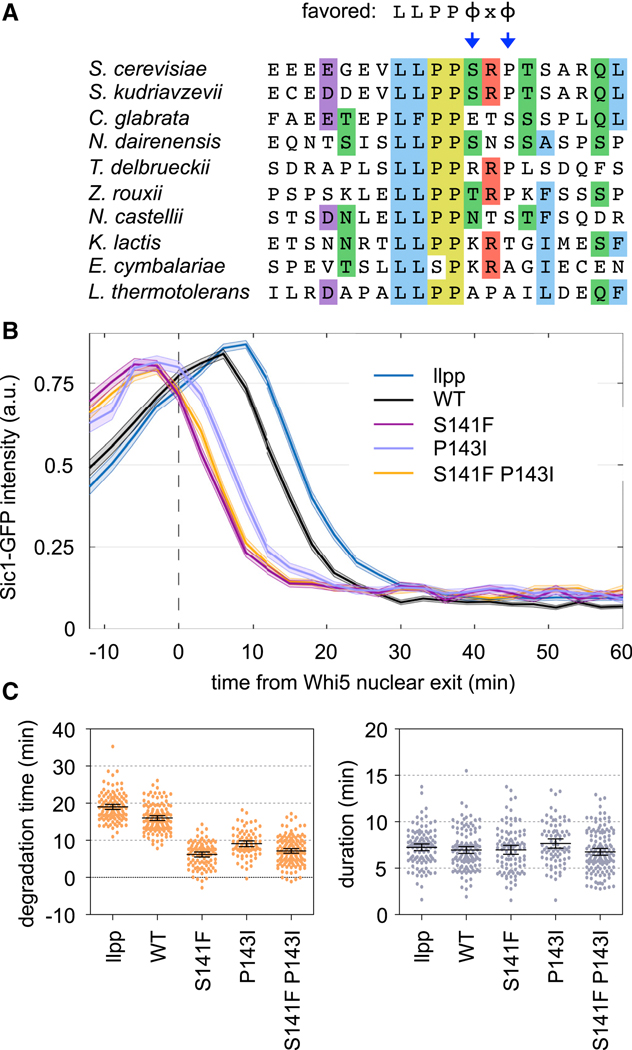
Figure 7. A weak Sic1 motif delays its CDK-mediated degradation.
(A) Sic1 LP sequences from ten budding yeasts; residues are colored where ≥ 50% are identical or similar. Preferred residues are at top; arrows emphasize the conserved absence of favorites at p5 and p7.
(B) Fluorescence intensities of Sic1-GFP variants, relative to the time when nuclear exit of Whi5-mCherry is 50% complete. Dark lines, mean; shaded bands, SEM. The “llpp” mutant substitutes VLLPP with AAAAA.
(C) Degradation time and duration (see Methods) for Sic1-GFP variants. Circles, individual cells; lines, mean and 95% CI.