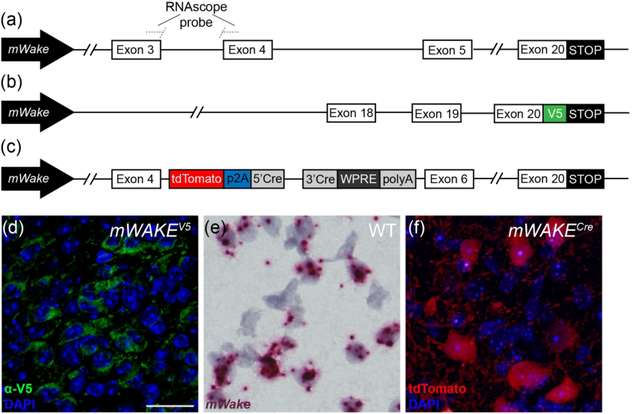
FIGURE 1. Methods for visualizing mWake expression.
The three main approaches for characterizing mWake expression are shown. (a) mWake mRNA was visualized using RNAscope, a high sensitivity technique for ISH, using probes bridging exons 3 and 4. (b) mWAKE protein was characterized by anti-V5 immunostaining of a transgenic mouse line where an inserted V5 epitope tag was fused to the C-terminus of mWAKE. (c) mWAKE+ cell bodies and processes were labeled by imaging tdTomato native fluorescence in a previously described transgenic mouse line (Bell et al., 2020), where exon 5 was replaced with a cassette containing both tdTomato and Cre-recombinase. Representative images from each approach are depicted. (d) IF using α-V5 antibodies (green) in the SCN of an mWake(V5/+) mouse shows mWAKE-V5 fusion protein in the cytosol. (e) Chromogenic labeling of mWake mRNA (red) via RNAscope ISH in wild-type SCN. (f) SCN neurons filled with tdTomato fluorescence (red) in an mWake(Cre/+) mouse, labelling both the cell bodies as well as the mWAKE+ processes. Images (d) and (f) also include the nuclear counterstain DAPI (blue), while (e) includes a hematoxylin counterstain (purple). The scale bar in (d) is 25 μm and applies to all images.