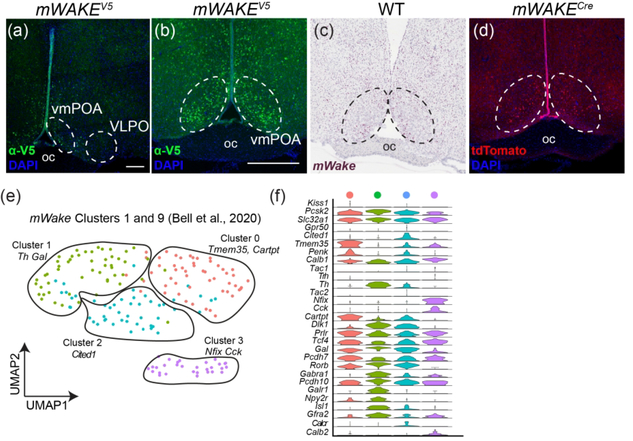
FIGURE 11. mWAKE+ cells and scRNA-Seq populations in the POA.
Representative images of mWake expression in the POA are shown. (a, b) Anti-V5 IF staining (green) with DAPI in mWake(V5/+) mice. (c) Chromogenic RNAscope ISH for mWake mRNA (red) with hematoxylin counterstain (purple) in wild-type (WT) mice. (d) Endogenous tdTomato fluorescence (red) in mWake(Cre/+) mice with DAPI. In (b-d), the dashed outlines denote the ventromedial POA. In (a), the scale bar denotes 200 μm. The scale bar in (b) is equivalent to 500 μm and applies to (c) and (d). e) UMAP plot of mWakePOA neurons demonstrating 4 sub-clusters marked by expression of key genes: cluster 0 (Tmem35, Cartpt, red, n=52 cells), cluster 1 (Th, Gal, yellow, n=51 cells), cluster 2 (Cited1, blue, n=49 cells) cluster 3 (Nfix, Cck, purple, n=30 cells). (f) Violin plot depicting the expression of key molecular markers for the clusters shown in (e). Each row depicts the expression of a single gene, with expression level on the y-axis, and the percentage of the cells expressing at that level forming the width. Abbreviations: oc, optic chiasm; VLPO, ventrolateral preoptic nucleus; vmPOA, ventromedial preoptic area.