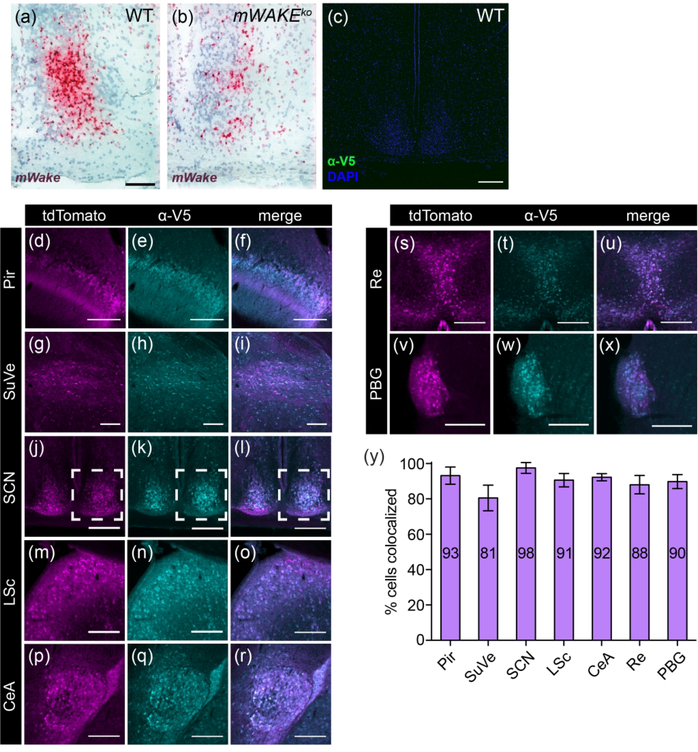
FIGURE 2. Validation of tools used to study mWake expression.
(a) Representative chromogenic RNAscope ISH for mWake mRNA (red) in SCN with hematoxylin counterstain (purple) in (a) wild-type (WT) (n=3) vs (b) mWake mutant (mWakeko) (n=3). The mWake mutant is described in Bell et al., 2020. In (a), the scale bar denotes 100 μm and also applies to (b). (c) Anti-V5 IF staining (green) of SCN with DAPI in a wild-type mouse, demonstrating negligible green signal. Scale bar in (c) denotes 200 μm. In (d-y), images representative of 7 locations with discrete mWake expression were used to assess correlation at the cellular level between mWakeV5 and mWakeCre transgenic models. Representative images from Pir (d-f), SuVe (g-i), SCN (j-l), LSc (m-o), CeA (p-r), cells adjacent to the Re (s-u), and PBG (v-x) are shown. Dashed outline in (j-l) indicates the unilateral portion used for quantification. (d), (g), (j), (m), (p), (s) and (v) Endogenous tdTomato fluorescence (magenta) in mWake(V5/Cre) mice. (e), (h), (k), (n), (q), (t), and (w) Anti-V5 IF staining (cyan) in the same mWake(V5/Cre) mice. (f), (i), (l), (o), (r), (u), and (x) Merged images for counting colabeled cells (white). (y) Colabeling quantification presented as % of V5+ cells with tdTomato fluorescence. Number of tdTomato+ cells and V5+ cells counted: Pir (n=404; n=423), SuVe (n=251; n=281), SCN (n=428; n=408), LSc (n=429; n=467), CeA (n=431; n=449), near Re (n = 430; n=468), PBG (n=246; n=261). Scale bars in (d-x) represent 200 μm. Abbreviations: CeA, central amygdala; LSc, lateral septal center; PBG, parabigeminal nucleus; Pir, piriform cortex; Re, thalamic reuniens nucleus; SCN, suprachiasmatic nucleus; SuVe, superior vestibular nucleus.