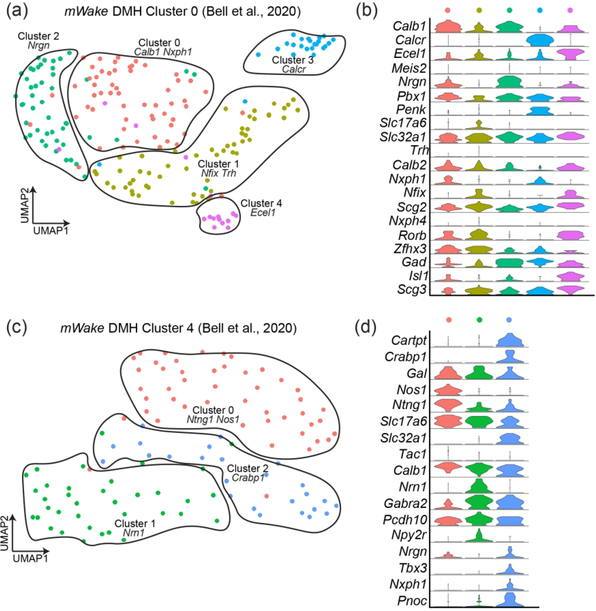
FIGURE 9. mWake+ scRNAseq populations in the DMH.
(a) and (c) UMAP plots of two mWakeDMH populations, with 5 and 3 subclusters, respectively. (b) and (d) Violin plots showing expression of key molecular markers for the subclusters shown in (a) and (c), respectively. Each row depicts the expression of a single gene, with expression level on the y-axis, and the percentage of the cells expressing at that level forming the width. For (a) and (b), the original population was named ‘Cluster 0’ in Bell et. al (2020), and the 5 subclusters are defined as cluster 0 (Calb1, Nxph1 red, n=57 cells), cluster 1 (Nfix, Trh, yellow, n=55 cells), cluster 2 (Nrgn, green, n=48 cells), cluster 3 (Calcr, blue, n=24 cells), cluster 4 (Ecel1, purple, n=16 cells). For (c) and (d), the original population was called ‘Cluster 4’ in Bell et. al (2020), and the 3 subclusters are defined as cluster 0 (Ntng1, Nos1, red, n=47 cells), cluster 1 (Nrn1, green, n=36 cells), cluster 2 (Crabp1, blue, n=25 cells).