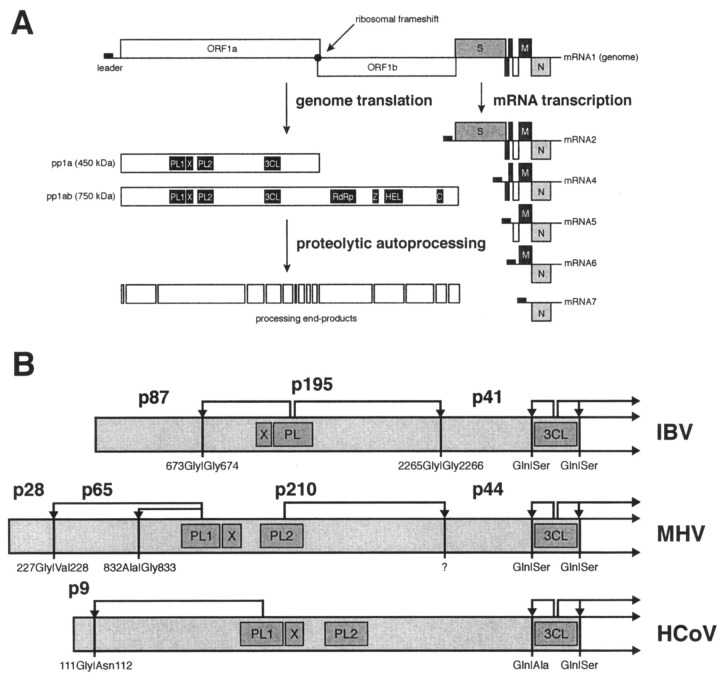
Figure 1.
Outline of theHCoVlife cycle and proteolytic processing of the N-terminal regions of coronavirus replicative polyproteins.A, ORFs in the polycistronic genome are indicated as boxes. The replicase gene, encompassing ORFs 1a and 1b, the gene for the surface glycoprotein protein, S, the triple-spanning membrane protein,M, and the nucleocapsid protein, N, are shown. The filled rectangle at the 5′ end of the genome represents the common leader sequence that is also present at the 5′ end of the subgenomic mRNAs that are shown below the genome. The conserved domains/functions encoded by the replicase gene are shown in the boxes depicting the two replicative polyproteins (pp1a and pp1ab). B, the N-terminal regions of the IBV, MHV, and HCoV replicative polyproteins pp1a/pp1ab are shown with the previously identified processing products and the corresponding cleavage sites (P1 and P1′ residues indicated). The following abbreviations are used:PL,papain-like protease; PL1, papain-like protease 1; X, domain conserved in coronaviruses, alphaviruses, rubiviruses, and hepatitis E virus (56); PL2, papain-like protease 2; 3CL, 3C-like protease;RdRp, RNA-dependent RNA polymerase;Z, putative zinc finger; HEL, NTPase/RNA helicase; C, conserved domain specific for nidoviruses (4).