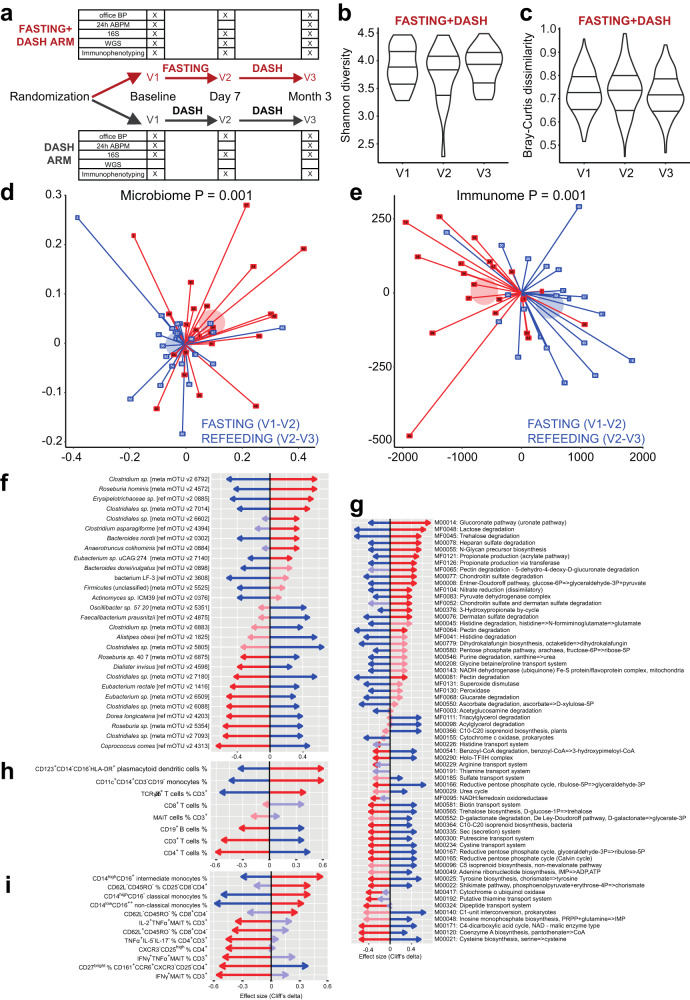
Fig. 1. Fasting has a pervasive host and microbiome impact.
a Study design is shown. Subjects are followed from baseline (V1), randomly assigned to begin a modified DASH diet only or to undergo a 5-day fast followed by a modified DASH diet. Follow-up is done at one week (V2) and 3 months (V3). b Fasting has no significant (two-sided MWU P > 0.05) impact on gut microbiome alpha diversity (Shannon diversity from mOTUv2 OTUs) across observation times V1–V3. c Fasting has no significant (two-sided MWU P > 0.05) impact on gut microbiome beta diversity (Bray–Curtis dissimilarity from mOTUv2 OTUs, shown are all between donor comparisons per time point) across observation times V1–V3. d Fasting significantly shifts the gut microbiome towards a characteristic compositional state, while refeeding reverses this change. Unconstrained Principal Coordinates graph with first two dimensions shown. Axes show Bray–Curtis dissimilarities of rarefied mOTUv2 OTUs between samples; each participant in the fasting arm is shown as two lines, one red (fasting change), one blue (refeeding change) connected (centered) at the origin for ease of visualization. Axes show fasting and refeeding deltas after one-week intervention and 3-month refeeding. Pseudonym participant ID numbers are shown on the point markers. Transparent circle markers show arithmetic mean position of fasting and recovery deltas, respectively. PERMANOVA test P-values reveal significant dissimilarity (P < 0.05) between samples from each visit V1–V3 in the original distance space, stratifying by donor. e Fasting significantly shifts the host immune cell population towards a characteristic state, while refeeding reverses it. Same as in (d), using Euclidean distances. f Gut microbial taxa significantly enriched/depleted upon fasting/refeeding. Taxa (mOTUv2 OTUs) are shown on the vertical axis, and effect sizes (Cliff’s delta) shown on the horizontal axis. Red arrows represent fasting effects (V2–V1 comparison), blue arrows refeeding effects (V3–V2 comparison). Bold arrows are significant (nested model comparison of a linear model for rarefied abundance of each taxon, comparing a model incorporating patient ID, age, sex and all dosages of relevant medications) to a model additionally incorporating time point, requiring likelihood test Benjamini-Hochberg corrected FDR < 0.1 and additionally pairwise post-hoc two-sided MWU test P < 0.05. g Gut microbial gene functional modules (KEGG and GMM models analyzed together) significantly enriched/depleted upon fasting/refeeding. h General immune cell populations significantly enriched/depleted upon fasting/refeeding. i Specific immune cell subpopulations. g–i Same test as in (f), subset of altered features shown for clarity. Effect sizes and FDR-corrected P values can be found in Supplementary Data 1,2.