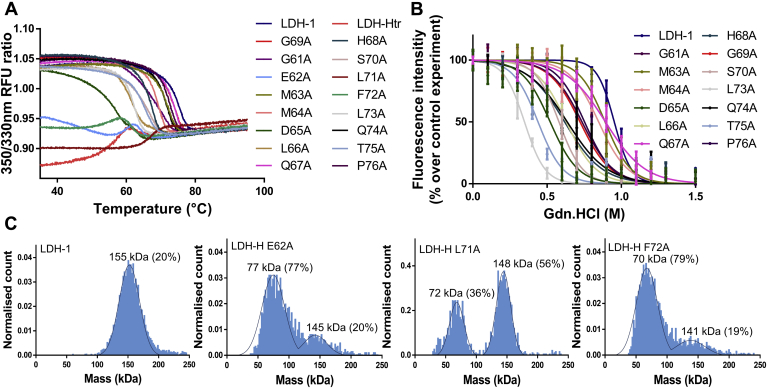
Figure 6.
Mutations of cluster B1unravel key residues for LDH tetramerization.A, screening of LDH-H variants using nanoDSF. Changes in the 350/330 nm fluorescence emission indicate blue or red shifts and are representative of unfolding events (n = 6). B, dissociation of the homotetrameric form of LDH-H and of its variants at 50 μg/ml (1.3 μM) upon addition of guanidinium·hydrochloride. Tryptophan fluorescence intensity was followed at λexc = 286 nm and λem = 350 nm as a direct reporter of LDH-1 tetrameric integrity (n = 6) (49). C, mass photometry was performed for different LDH-H variants with the experimental molecular weights of the complexes in solution and their relative intensities. Theoretical molecular weight of the tetramer = 155 kDa; theoretical molecular weight of the dimer = 78 kDa. Profiles of the other variants can be found in Fig. S4. λem, wavelength of emission; λexc, wavelength of excitation; LDH, lactate dehydrogenase; LDH-1, lactate dehydrogenase heart isozyme homotetramer; LDH-H, lactate dehydrogenase heart isozyme; nanoDSF, nanoscale differential scanning fluorimetry; RFU, relative fluorescent unit.