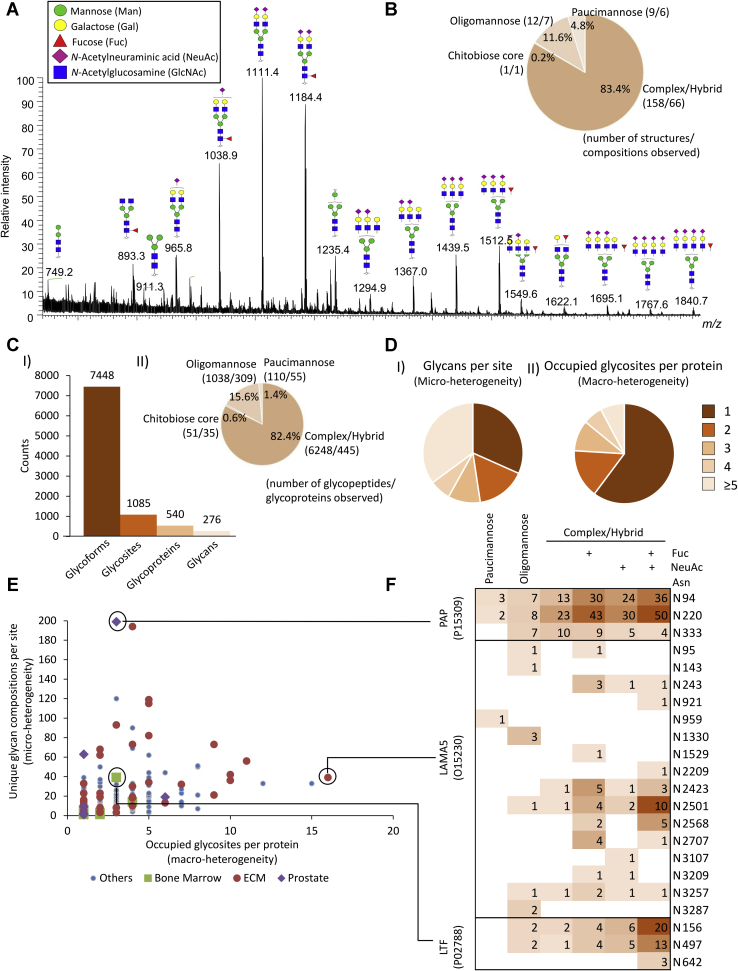
Fig. 2.
The prostate cancer (PCa) tissue N-glycome and N-glycoproteome.A, summed MS spectrum (10–68 min) from a representative LC–MS/MS N-glycan profile of PCa tissue (G1). B, N-glycan class distribution based on the identified glycans across the investigated PCa grades and BPH (average of the relative abundance from all samples). C, (I) overview of the N-glycoproteome of PCa tissue including the identified unique N-glycoforms (unique protein + unique site + unique glycan), N-glycosites, N-glycoproteins, and N-glycans identified in ZIC-HILIC-enriched fractions. (II) N-glycan class distribution based on identified intact glycopeptides. D, microheterogeneity (I) and macroheterogeneity (II) of protein N-glycosylation based on the glycans (unique) observed at each site and the occupied glycosites per protein. E, glycan heterogeneity by protein. The cell source of the identified proteins has been annotated according to the Protein Atlas, see key for color coding. F, extensive microheterogeneity and macroheterogeneity of selected glycoproteins from different cell origins including the prostate tissue–derived prostatic acid phosphatase (PAP), ECM-derived laminin subunit alpha-5 (LAMA5), and bone marrow–derived lactotransferrin (LTF) in the PCa tumor microenvironment. ECM, extracellular matrix.