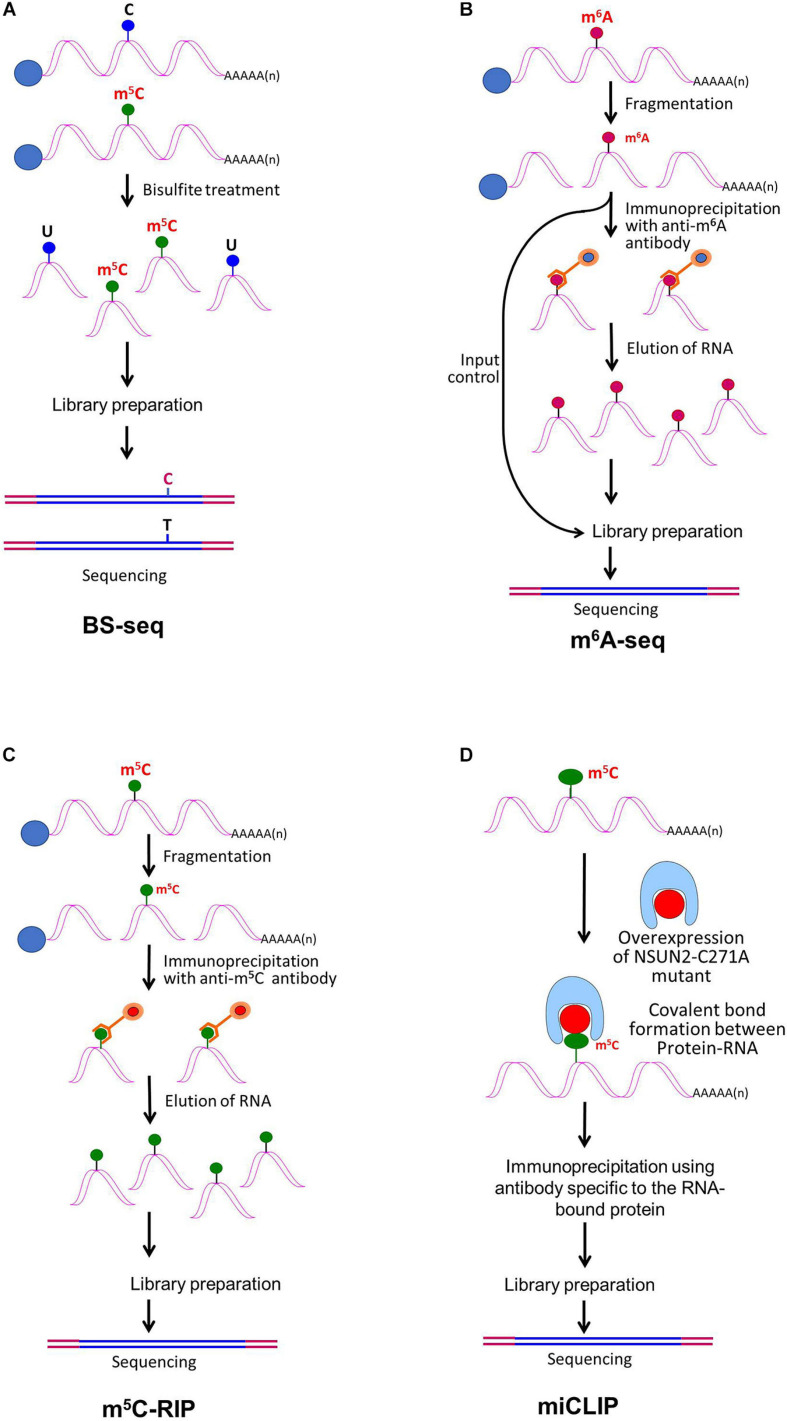
FIGURE 3.
Detection of modified bases in mRNA. (A) Bisulfite sequencing (BS-seq) for the detection of 5-methylcytosine (m5C). Purified mRNA is fragmented into small (100–200 nt) fragments, and subjected to bisulfite treatment. Bisulfite treatment causes converts cytosine (C) to uracil (U), but m5C remains unchanged. Presence of C is detected by sequencing, wherein it is replaced by T. (B) Purified mRNAs are fragmented into 100–200 nt, followed by immunoprecipitation using anti-m6A antibody to enrich the sample with fragments containing the modified base, library preparation, and high-throughput deep-sequencing for detection of m6A. (C) Purified mRNAs are fragmented followed by immunoprecipitation using anti-m5C antibody of the fragments containing the modified base, library preparation, and sequencing. (D) m5C individual-nucleotide-resolution crosslinking and immunoprecipitation (m5C-miCLIP) exploites catalytic activity of cysteine-to-alanine mutation (C271A) mutant of NSUN2 (methyltransferase) which inhibits release of the enzyme from the protein–RNA complex making stable covalent bond between NSun2 and its RNA targets. Antibody specific to the RNA bound protein is used for immunoprecipitation, followed by library preparation and sequencing. This allows detection of low-abundance methylated RNAs without the need of deep sequencing.