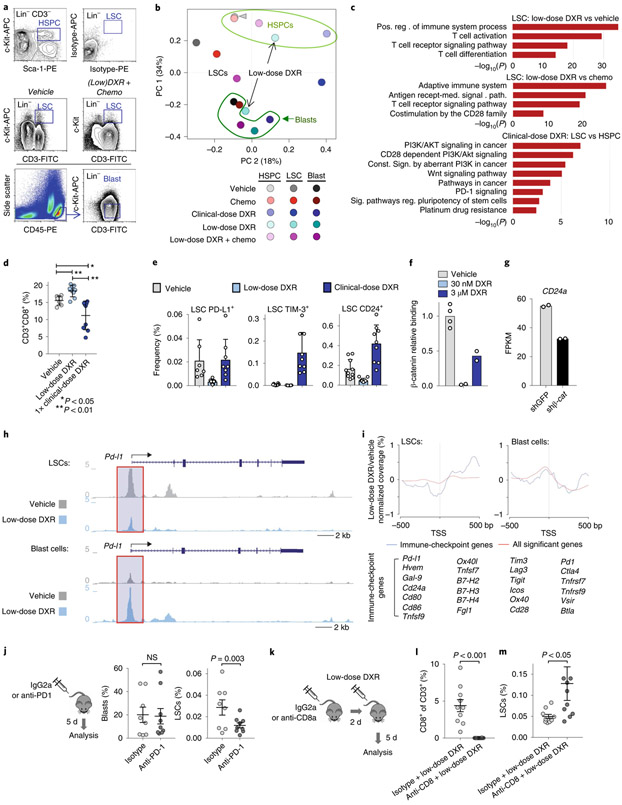
Fig. 4 ∣. LSCs uniquely express immune checkpoints that can be inhibited with low-dose DXR.
a, Transcriptome analysis of indicated populations sorted by FACS at 5 d after treatment; representative gates are indicated. To obtain sufficient cells from the clinical DXR sample, the 5 × 4 mgkg−1 DXR daily dosing schedule was used. At least one replicate experiment yielded similar results for each. b, Principal component analysis (arrowhead indicates overlap of HSPCs from vehicle and chemotherapy). c, Gene ontology enrichment analysis using −log10 of the uncorrected P value as the x axis. Upregulated enriched terms are shown for the indicated population and treatment comparisons. In b,c, n = 2 biologically independent replicates for each (see Methods). d, Peripheral blood from leukaemic mice was analysed using FACS for CD8+ T cells at 48 h after treatment with vehicle, low-dose DXR or clinical DXR (1 × 20 mgkg−1). n = 7 (vehicle), 9 (low-dose DXR) and 9 (clinical DXR) biologically independent mice; data are mean ± s.d. e, LSCs were analysed 24–72 h after treatment for the indicated immune checkpoints. n = 7 biologically independent mice for each; mean ± s.d. f, Relative binding of β-catenin to the Cd24a gene locus (fold enrichment by ChIP-qPCR, normalized to the vehicle samples) 4 h after low (30 nM) or high (3 μM) DXR treatment. n = 4 (vehicle), 2 (30 nm DXR) and 2 (3 μM DXR). g, Cd24a expression in mouse ES cells with β-catenin knockdown (shβ-cat) for 48 h compared with the control (shGFP). n = 2 independent experiments each. FPKM, fragments per kilobase of transcript per million mapped reads. h, Chromatin accessibility profiles of example immune-checkpoint genes observed by ATAC-seq in LSCs and blast cells (see Methods). i, Metagene analysis of LSCs showing chromatin accessibility near the transcriptional start site (TSS) in immune-checkpoint gene loci (listed) compared with all loci showing significant changes. Gene synonyms are listed here in parentheses: Hvem (Tnfrsf14), Gal-9 (Lgals9), Ox40l (Tnfsf4), Tnfsf7 (Cd70), B7-H2 (Icosl), B7-H3 (Cd276), B7-H4 (Vtcn1), Pd1 (Pdcd1), Tnfrsf7 (Cd27), Tim3 (Havcr2), Ox40 (Tnfrsf4). j, Leukaemic mice were treated with PD1 antibody or isotype control and analysed by FACS. n = 8 (isotype) and 9 (anti-PD1) biologically independent mice; data are mean ± s.e.m. k–m, CD8+ T cells were depleted with CD8 antibody before low-dose DXR treatment; bone marrow was analysed by FACS. n = 10 biologically independent mice for each group; data are mean ± s.e.m. In d,j,l,m, unpaired two-tailed t-test.