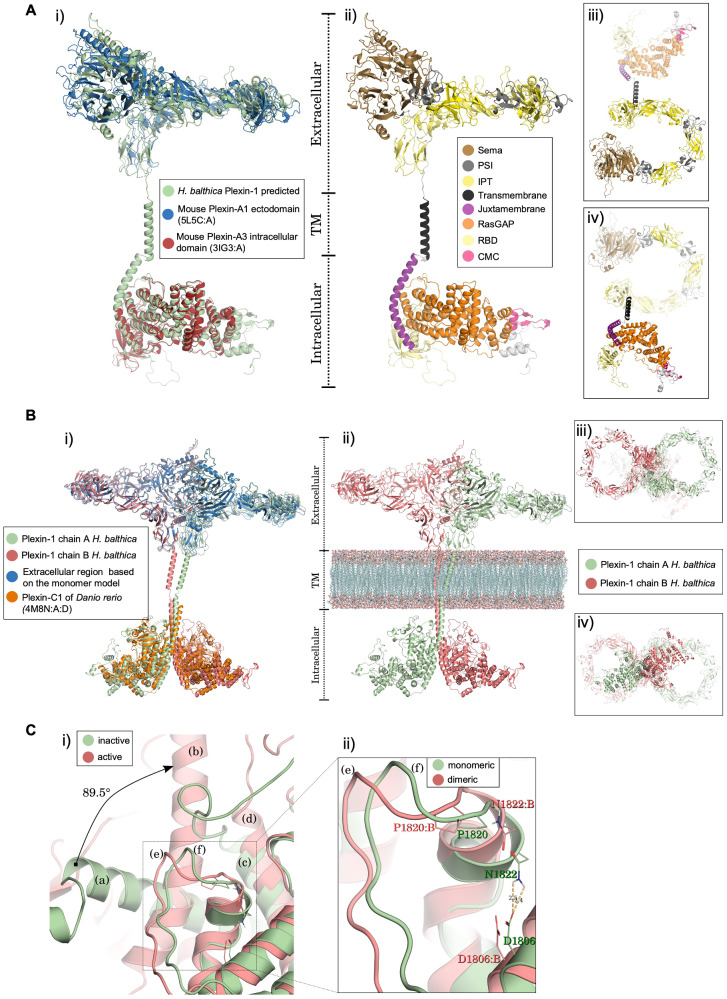
Fig. 4.
Structural 3D models of choanoflagellate Plexin-1. (A) Monomer model of H. balthica (Hbalt) Plexin-1. (i) Structural alignment of monomeric Hbalt Plexin-1 with the ectodomain of mouse Plexin-A1 (PDB 5L5C: A) and the intracellular domain of mouse Plexin-A3 (3IG3: A). (ii–iv) Domain organization of Hbalt Plexin-1, shown as front view (ii), top view (iii), and bottom view (iv). Extracellular domains: SEMA, PSI, and IPT. Transmembrane region: TM. Intracellular domains: JM (juxtamembrane helix), Ras-GAP, RBD, and CMC (conserved motif near C-terminus). (B) 3D model of dimeric Hbalt Plexin-1. (i) Structure of Hbalt Plexin-1 dimer model (chains A and B) aligned with extracellular region of monomer model and intracellular regions of dimeric zebrafish Plexin-C1 (4M8N: A: D). (ii) Overview of extracellular, transmembrane, and intracellular domains (front view): chains A and chain B traversing model membrane (phosphatidylcholine) constructed with VMD software. (iii) Top view of the dimeric model. (iv) Bottom view of the dimeric model. (C) Structural changes predicted upon dimerization of Hbalt Plexin-1. (i) Comparison between the inactive (monomeric) and active (after dimerization) structures. The juxtamembrane helix rotates 89.5° from the inactive (a) to the active structure (b) (chain A). The helix 11 (according to Wang et al., 2013) remains stable in both conformations (c and d). The activation segments (e) and (f) also undergo conformational changes. (ii) Detailed view of conformational changes of activation segment between monomeric and dimeric structures. The displacement in the activation segment of dimeric conformation (chain B) promotes breakdown of the hydrogen bond between D1806: B and N1822: B.