Fig. 6.
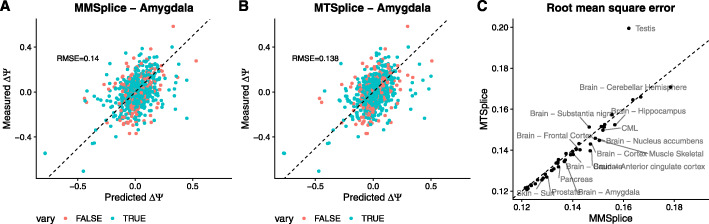
Comparing MMSplice and MTSplice on predicting variant-associated differential splicing. a, b Predicted (x-axis) versus measured (y-axis) ΔΨ in amygdala between alternative and reference alleles for variants with between-tissue splicing variation (orange) and other variants (cyan) for MMSplice (a) and MTSplice (b). c Root-mean-square error of MTSplice predictions (y-axis) against MMSplice predictions (x-axis) for exons with between-tissue splicing variations (the cyan dots in a). Each dot represents one of the 51 GTEx tissues with at least 10 measured variant effects. MTSplice improves for 39 tissues, yet mildly, over MMSplice. Tissues for which the RMSE differences larger than 0.002 are labeled with text