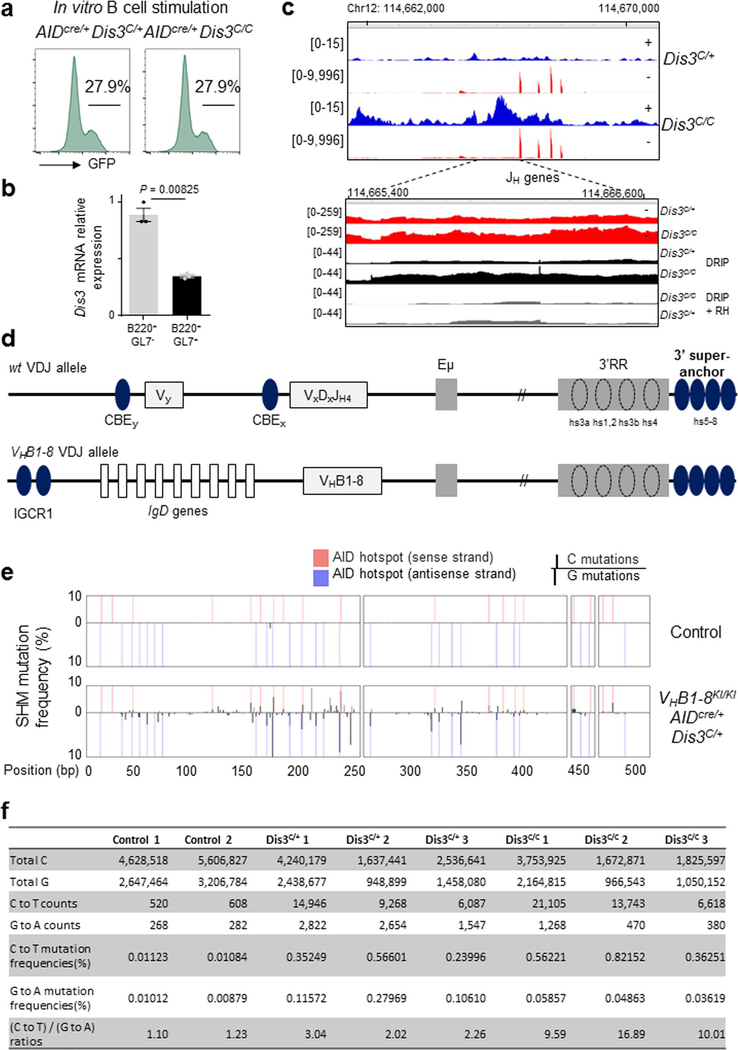
Extended Data Fig. 10 |. In vivo somatic hypermutation exploration in Dis3C/C B cells.
a. B splenocytes from one pair of AIDcre/+ Dis3C/+ and AIDcre/+ Dis3C/C mice were stimulated. GFP expression was examined after 4 days. In ex vivo culture conditions AIDcre/+ Dis3C/C cells do not encounter selection and maintain numbers of GFP+ cells. b. RT-qPCR quantification of Dis3 mRNA expression in AIDcre/+ Dis3C/C Peyer’s patches B cells. B220+ GL7+ and B220+ GL7− cells were sorted to show the specific deletion of the Dis3COIN allele in activated B cells (one experiment performed in triplicate, mean is shown +/− s.e.m., two-tailed unpaired t-test). c. RNA-seq showing increased ncRNAs overlapping JH genes in the absence of DIS3 activity during B cell stimulation. Top. RNA-seq tracks encompassing the Eμ intronic enhancer and JH genes. Bottom. RNA-seq tracks showing increased sense transcription (red) and DNA:RNA hybrid accumulation (black) downstream of the JH4 gene in the absence of DIS3 activity (2 RNA-sequencing and 3 DRIP-sequencing) d. Top: Schematic of wt recombined VDJ genes in the context of CBEs. Bottom: VHB1–8KI alleles have different configuration, conserving the major CTCF-binding region IGCR1 (IGCR1 is deleted from the functional allele in physiological conditions). e. DNA mutation analyses of GC (B220+ GL7+) B cells from VHB1–8KI/KI AIDcre/+ Dis3C/+ (n = 3) mice. Tail DNA from the same animals was used as control (n = 2). AID hot-spots are indicated, and C or G mutations refer to the sense DNA strand. f. Quantification of C and G mutation frequencies in GC B cells. 3 pairs of VHB1–8KI/KI AIDcre/+ Dis3C/+ and VHB1–8KI/KI AIDcre/+ Dis3C/C mice were used, analyses were performed at the JH gene from the VHB1–8 allele. χ2 two-tailed proportions tests were used to compare the number of C to T mutations relatively to the total C sequenced, and the number of G to A mutations compared to the total G sequenced.