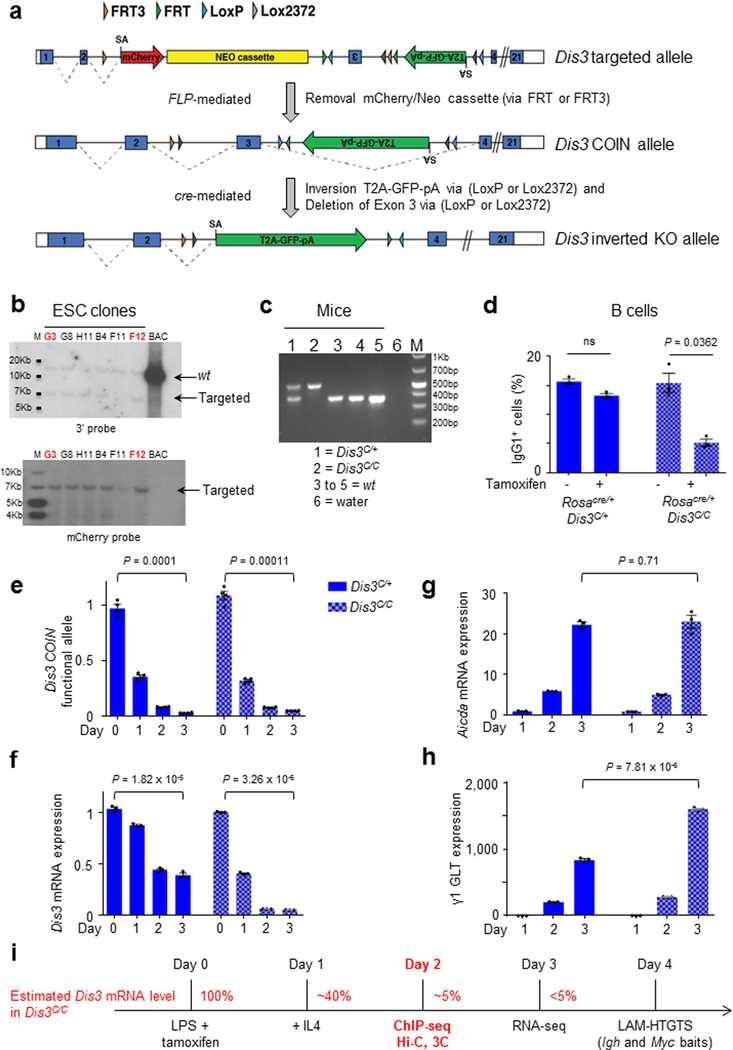
Extended Data Fig. 1 |. Generation of Dis3COIN mouse model and efficient Dis3 deletion in activated B cells.
a, Dis3 targeted allele includes mCherry and Neo resistance cassettes, inverted Dis3 exon 3, GFP gene, and loxP/FRT sites (top). After FLP-mediated removal of mCherry/Neo cassettes and exon 3 reversion, Dis3COIN allele is functional (middle). Dis3 KO allele after cre-mediated inversion (bottom). b, ESC screening by Southern blot with 3′ or mCherry probes. Clones G3 and F12 were selected for microinjection. Screening was performed one time, each clone tested with different probes. c, Mouse genotyping. Dis3 wt allele generates a band of 369 bp, Dis3COIN allele 489 bp. This gel is representative of our genotyping, performed at least 10 times. d, Rosacre/+ Dis3C/+ and Rosacre/+ Dis3C/C B cells were stimulated in the presence (+) or in the absence (−) of tamoxifen, and CSR to IgG1 was quantified (3 independent experiments, mean is shown +/− s.e.m., two-tailed paired t-test). e, qPCR quantification of Dis3 allelic inversion on genomic DNA, normalized to the GFP gene. This qPCR is specific of the Dis3 COIN allele before inversion, and shows loss of the Dis3 functional allele after inversion. f, g, and h, RT-qPCR quantification of Dis3, Aicda (AID) mRNAs, and γ1 germline transcript expression on total RNA, normalized to Gapdh. Kinetic in panels e to h was performed one time in triplicate, mean is shown +/− s.e.m., two-tailed unpaired t-test). i, Resting B splenocytes were collected and stimulated with LPS and tamoxifen to inactivate Dis3COIN alleles, IL4 was added at day 1. ChIP, Hi-C, and 3C experiments were performed at early time points. RNA-seq were performed 2 days after IL4 treatment, while cells were harvested at late time-point to study the accumulated DNA junctions from CSR or translocations by LAM-HTGTS.