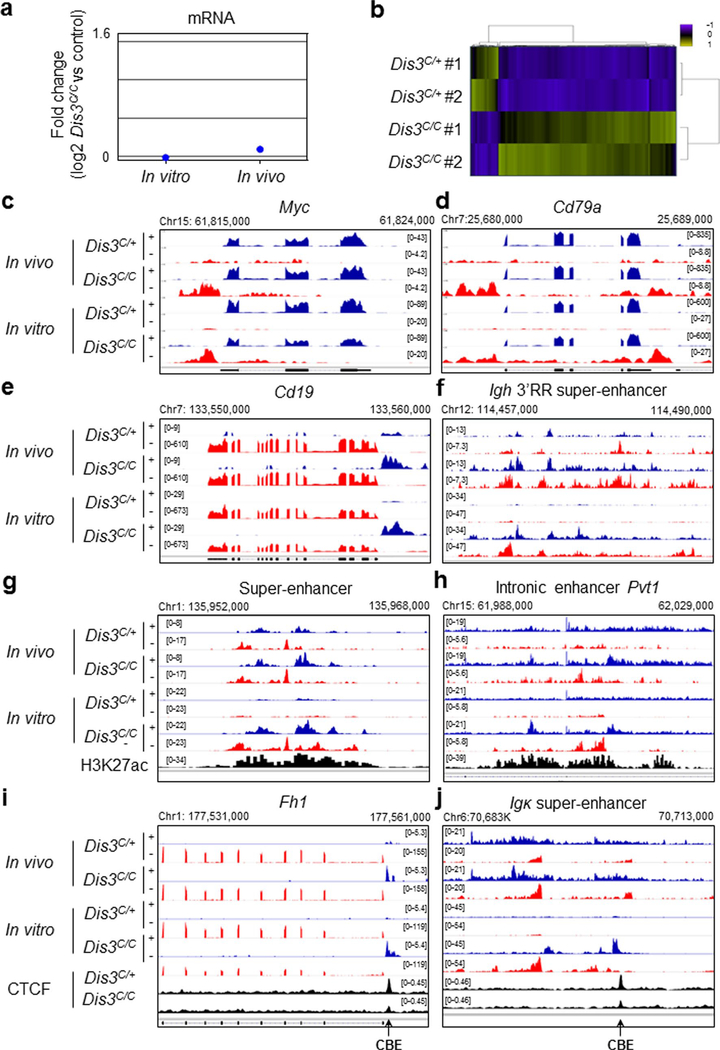
Extended Data Fig. 2 |. Accumulation of DIS3-sensitive ncRNAs in B cells isolated from the germinal center in vivo and stimulated in vitro.
a, mRNA expression in vitro and in vivo. RNA-sequencing from Rosacre/+ Dis3C/+ versus Rosacre/+ Dis3C/C in vitro activated B cells (n = 2), and in vivo AIDcre/+ Dis3C/+ versus AIDcre/+ Dis3C/C (n = 2) were analyzed. b, Differential gene expression in DIS3-deficient B cells. RNA-sequencing from Rosacre/+ Dis3C/+ and Rosacre/+ Dis3C/C activated B cells (n = 2) were analyzed using a hierarchical unsupervised clustering method and results are shown as heat maps. Up-regulated genes are shown in green and down-regulated genes are shown in blue. A total of 160 genes showed differential RNA expression at this early time-point. c to e, Examples of DIS3-sensitive aTSS and antisense RNAs at various genes, including Myc, Cd19, and Cd79a, genes frequently translocated in B cells. f, eRNAs expressed from the Igh 3′RR super-enhancer. g, Example of DIS3-sensitive enhancer RNAs (eRNAs) at super-enhancer, with the active enhancer mark H3K27ac. h, Example of DIS3-sensitive eRNAs at intronic enhancer, from Pvt1 locus, with the active enhancer mark H3K27ac. i, Example of DIS3-sensitive CBE-overlapping RNAs at Fh1 locus. j, Example of DIS3-sensitive CBE-overlapping RNAs downstream the Igk super-enchancer.