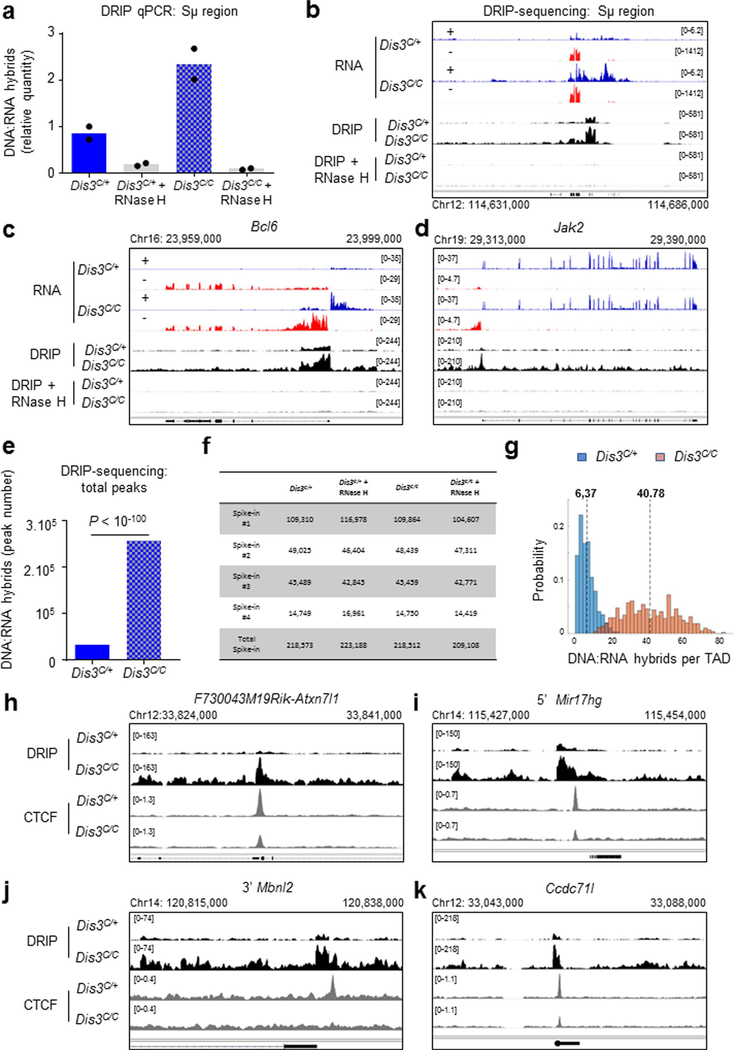
Extended Data Fig. 3 |. Accumulation of DNA-associated RNAs in the absence of DIS3.
a DNA:RNA immunoprecipitation (DRIP). DRIP was performed on Rosacre/+ Dis3C/+ and Rosacre/+ Dis3C/C activated B cells. DNA was extracted, digested by restriction enzymes, and treated with RNase H for the negative controls before immunoprecipitation using S9.6 antibody. qPCR quantification of DNA:RNA hybrids at Sμ region. DRIP products were quantified directly by qPCR, relative to the input. One experiment performed in duplicate, each dot represents one qPCR value. b to j, DRIP-sequencing: these DRIP products were prepared for deep sequencing. 3 deep-sequencings were performed with similar results. Coding and non-coding transcription determined by RNA-sequencing is shown (‘RNA’ tracks). b, DNA:RNA hybrids accumulation at Sμ region, relative to a. c. Example of DNA:RNA hybrids accumulation at the Bcl6 gene, correlating with the accumulation of intronic ncRNA in DIS3-deficient cells. d, Example of DNA:RNA hybrids accumulation at the Jak2 gene. e, Peak calling was applied to quantify the different peaks corresponding to DNA:RNA hybrids in DRIP sequencing experiments. Quantification is shown in Rosacre/+ Dis3C/+ and Rosacre/+ Dis3C/C activated B cells (bar graphs show the number of peaks, χ2 two-tailed proportions test). f, Read numbers from the different spike-in controls used in DRIP experiments are shown. g, Distribution of DNA:RNA hybrids per TAD in Rosacre/+ Dis3C/+ and Rosacre/+ Dis3C/C activated B cells, means are indicated by dashed lines. h to k, Multiple examples of accumulation of DNA:RNA hybrids overlapping CBEs and decreasing CTCF binding in the absence of DIS3.