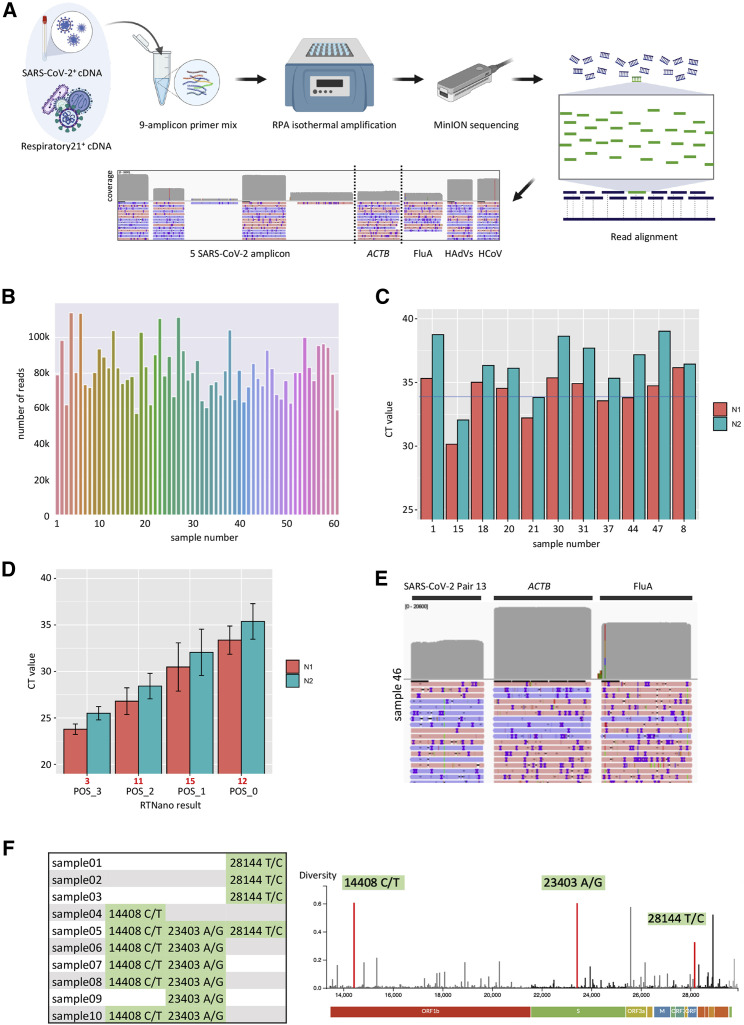
Figure 3.
Real-time detection of multiple viral pathogens and mutational analysis of SARS-CoV-2
(A) Experimental design of multiple virus detection by one-pot NIRVANA. A mixture of SARS-CoV-2+ and Respiratory21+ samples was used as positive control to adjust the primer concentration. The final primer mix could amplify all targeted viral regions. Created with BioRender.com.
(B) The sequencing throughput of 60 clinical samples (1–60) and NTC (61). A total of 6.3 million reads were acquired in a 24-h sequencing run.
(C) CT values of potentially false-negative samples by RTNano analysis. The average CT value of the N1 assay was indicated by the blue line.
(D) The average rRT-PCR CT values of SARS-CoV-2 RTNano+ samples (PCR+ of both N1 and N2 assays) of different confidence level using 9-amplicon NIRVANA. The sample number is shown in red under the graph. The error bars represent the standard deviation of CT values. RTNano confidence level inversely correlates with CT value.
(E) IGV plots showing the read alignment to the SARS-CoV-2, ACTB, and FluA amplicon in sample 46 using 9-amplicon NIRVANA.
(F) The SNVs detected in multiplex RPA sequencing and their position as shown in the Nextstrain data portal (https://nextstrain.org). A total of 16 SNVs were detected from 10 SARS-CoV-2-positive samples.