Extended Data Figure 5. The validation of candidate proteins with CRISPR-based astrocytic candidate gene depletion strategy.
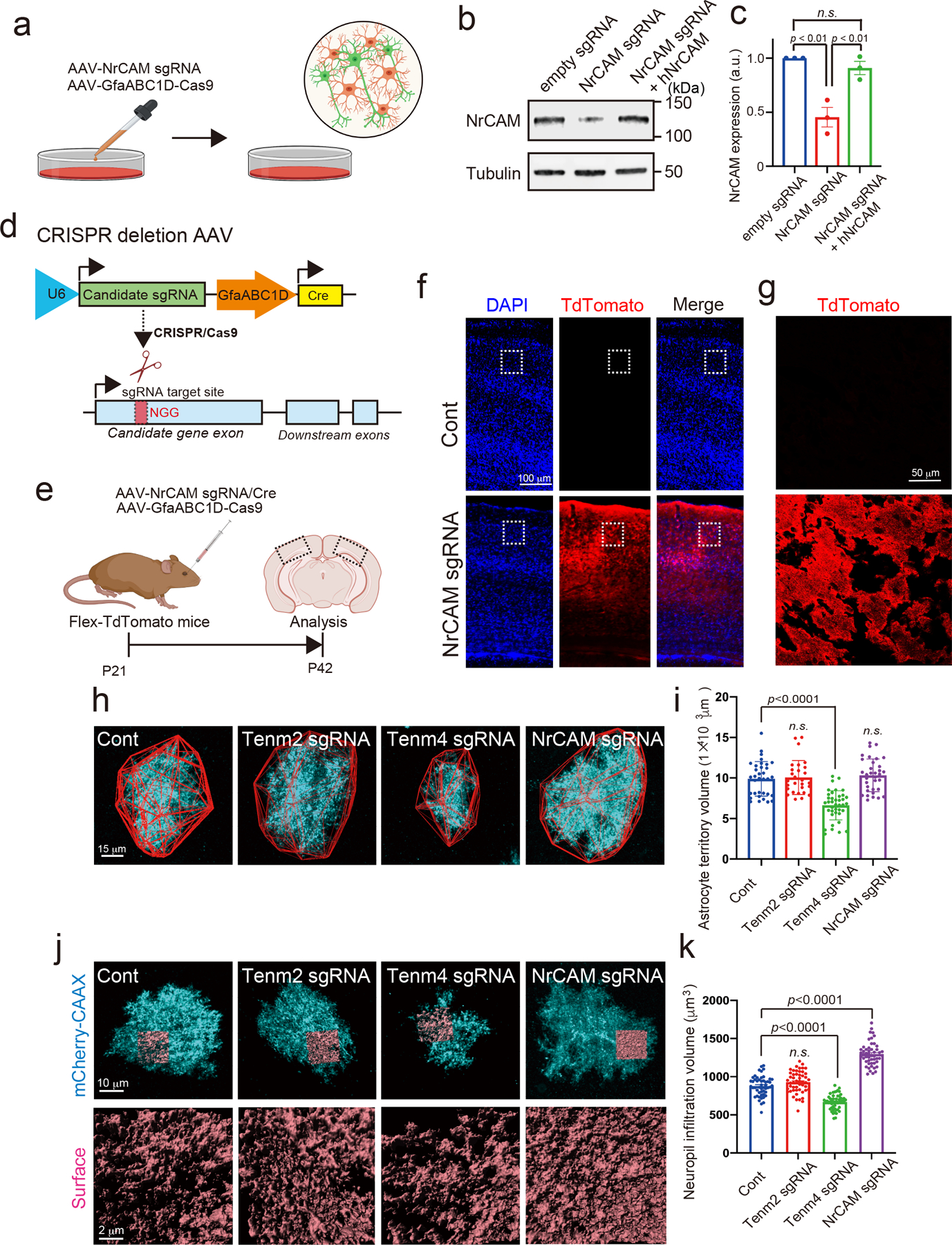
a, Schematic of CRISPR-based deletion of astrocytic NrCAM in vitro. b, Immunoblots showing loss of NrCAM with sgRNA. AAV1/2-U6-empty sgRNA or AAV1/2-U6-NrCAM sgRNA was co-infected with AAV1/2- GfaABC1D-Cas9 to cultured neurons and astrocytes at DIV14. The cells were subjected to immunoblot analysis with an anti-NrCAM antibody. Tubulin was used as a loading control. c, The bar graph indicates the expression level of NrCAM from 3 independent experiments. d, Schematic of CRISPR-based deletion strategy of candidate gene. e, Experimental timeline of AAV-mediated CRISPR-based astrocytic gene deletion strategy in Flex-TdTomato mice. f, AAV PHP.eB-U6-NrCAM sgRNA was co-infected with AAV PHP.eB-GfaABC1D-Cas9 in Flex-TdTomato mice at P21. Coronal sections were prepared and immunostained with an anti-TdTomato antibody. g, A High-magnification image is shown. h, Images of Tenm2-, Tenm4- or NrCAM-deleted astrocytes (cyan) and their territories (red outlines) in visual cortexes of juvenile mice. i, Average territory volumes at P42 of Tenm2-, Tenm4- or NrCAM-deleted astrocytes. Between 20–25 cells per condition from 3 mice. j, Images of Tenm2-, Tenm4- or NrCAM-deleted astrocytes (cyan) and their NIV reconstructions (orange) in visual cortexes of juvenile mice. k, Average NIV at P42 of Tenm2-, Tenm4- or NrCAM-deleted astrocytes. 51 cells per each condition from 3 mice. n= 3 biological repeats. One-way ANOVA (Dunnett’s multiple comparison, p<0.0001, 0.01). Data are means ± s.e.m.
