Figure 4.
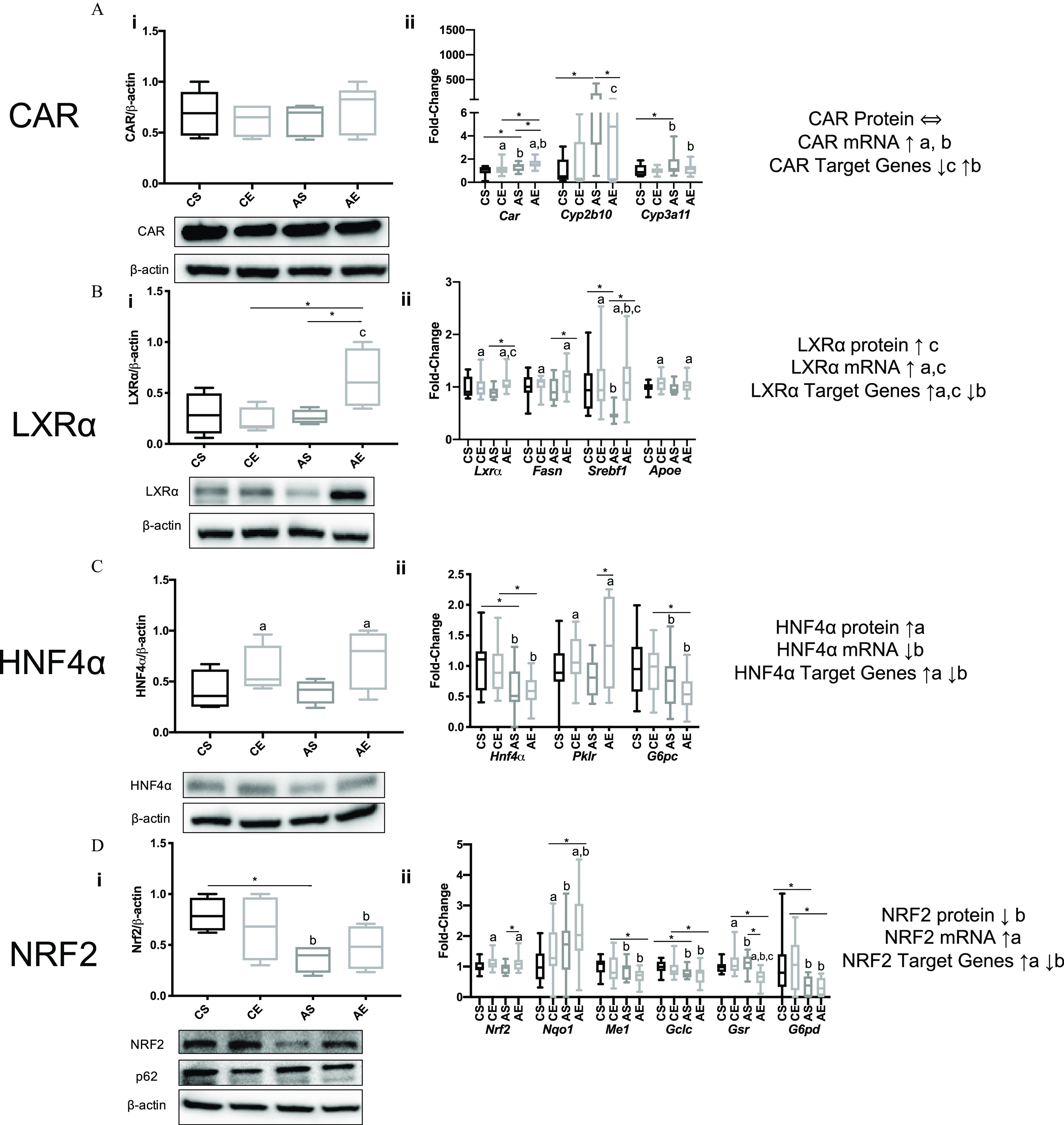
Expression of select hepatic nuclear receptors, AhR, and NRF2 and their respective target genes in HFD-fed mice exposed to vehicle or treatment. Male WT C57Bl/6 mice were fed an HFD (12 wk) Aroclor 1260 () by a one-time oral gavage at week 1. Aroclor 1260-exposed or vehicle mice were treated (via IP injection) with saline or EGF () daily for 10 d starting at week 10. Mice were fasted for 12 h and euthanized, and tissues were harvested for downstream analyses. (Ai–Di) Western blot analysis measuring hepatic (Ai) CAR, (Bi) , (Ci) , (Di) NRF2, and p62 protein expression in mice gavaged with corn oil and injected with either saline (CS) or EGF () (CE) or gavaged with Aroclor 1260 () and injected with saline (AS) or EGF (AE). (Aii–Dii) qPCR analysis measuring hepatic mRNA for (Aii) CAR and CAR/PXR target genes Cyp2b10/Cyp3a11, (Bii) and target genes Fasn, Srebf1, and Apoe, (Cii) and target genes Pklr, and G6pc (Dii) Nrf2 and target genes Nqo1, Me1, Gclc, Gsr, and G6pdh. An was used for the western blot analysis data, and an was used for all the qPCR data. A two-way ANOVA was used to statistically compare data sets followed by a Sidak correction for intergroup comparisons. A was considered significant. A is denoted by * for the Sidak multiple comparison test. Significance due to EGF is denoted by (a), due to Aroclor 1260 denoted by (b), and due to interaction denoted by (c) for the two-way ANOVA. Data are presented as box and whisker plots for Figure 4 that illustrate the median (midline), upper and lower quartiles (box), and the upper and lower limits (whiskers). All numerical data are presented in Table S6 “Numerical Data” as and p-values can be found in Table S5. Numeric outliers were identified by ROUT method and removed. The number of outliers are reported in Table S4. Note: AE, ; ANOVA, analysis of variance; Apoe, apolipoprotein E; AS, ; CAR, constitutive androstane receptor; CE, ; CS, ; Cyp2b10, cytochrome P450 2B10; Cyp3a11, cytochrome P450 3A11; EGF, epidermal growth factor; Fasn, fatty acid synthase; HFD, high-fat diet; , hepatocyte nuclear receptor 4-alpha; Gclc, glutamate-cysteine ligase catalytic subunit; Gsr, glutathione-disulfide reductase; G6pc, glucose-6-phosphatase; G6pdh, glucose-6-phosphate dehydrogenase; IP, intraperitoneal; , liver X receptor-alpha; NRF2, nuclear factor-erythroid 2-related factor 2; Me1, malic enzyme 1; Nqo1, NAD(P)H quinone dehydrogenase 1; Pklr, pyruvate kinase, liver type; PXR, pregnane xenobiotic receptor; p62, sequestosome 1; Srebf1, sterol regulatory element binding transcription factor 1.
