Figure 5.
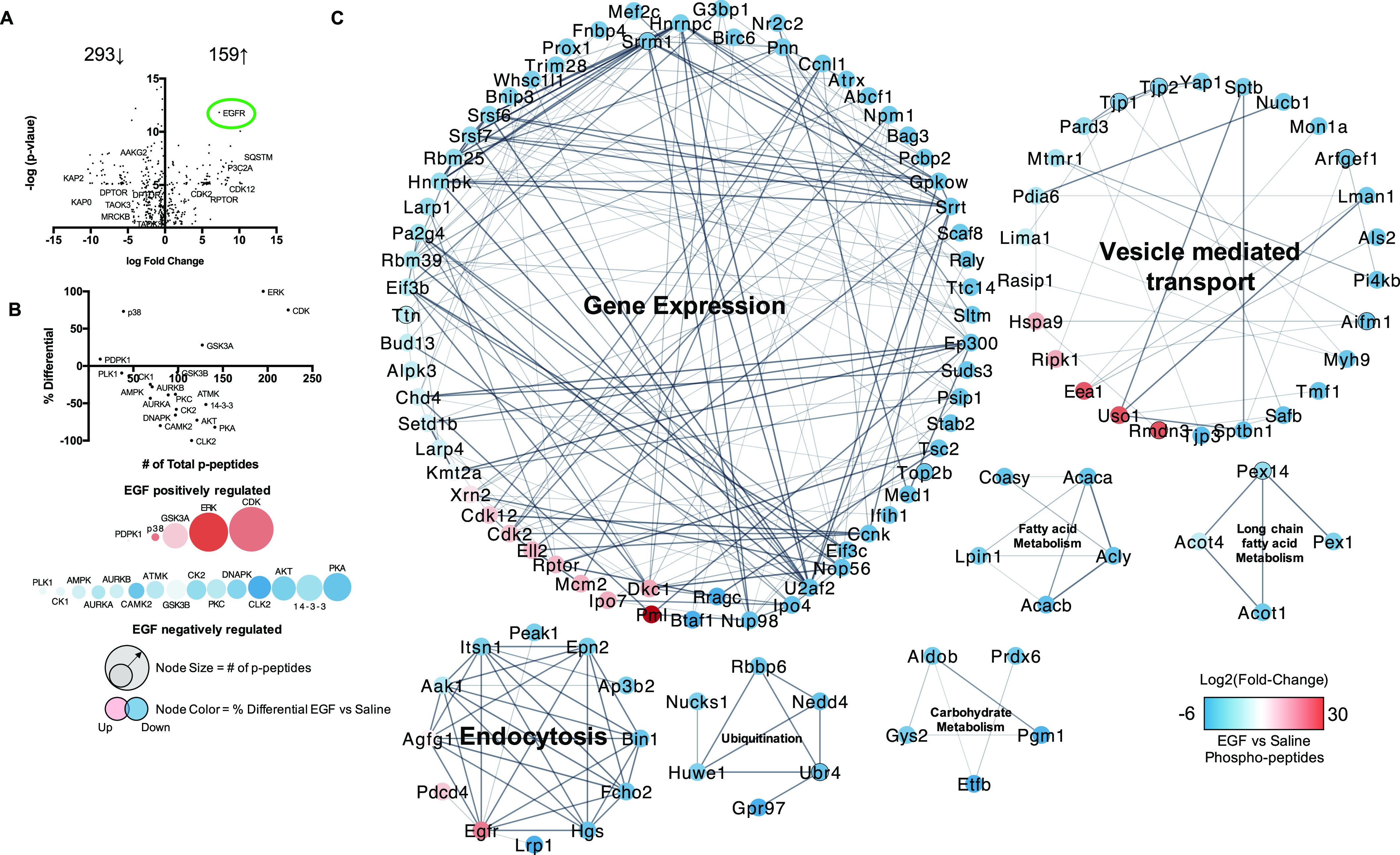
Characterization of the EGF-sensitive hepatic phosphoproteome and kinome: Male WT C57Bl/6 mice fed a control diet were treated with either saline or EGF (), followed by euthanasia at 30 min post IP injection and liver resection for downstream phosphoproteomic analysis. (A) Volcano plot of log 2(fold-change) and -log(p-value) for hepatic phosphopeptides (EGF vs. saline) significantly changed with EGF () IP after 30 min. (B) Kinome analysis based on phosphopeptides significantly regulated by EGF administration in the liver. (C) Cluster and GO processes analysis of the significant EGF-sensitive Phosphopeptides (Clusters numbered 1–7). An was used for the acute phosphoproteomic study, and a was considered significant by a two-tailed t-test. Pathways and processes had to meet an . Note: CE, ; CS, ; EGF, epidermal growth factor; FDR, false discovery rate; GO, gene ontology; IP, intraperitoneal.
