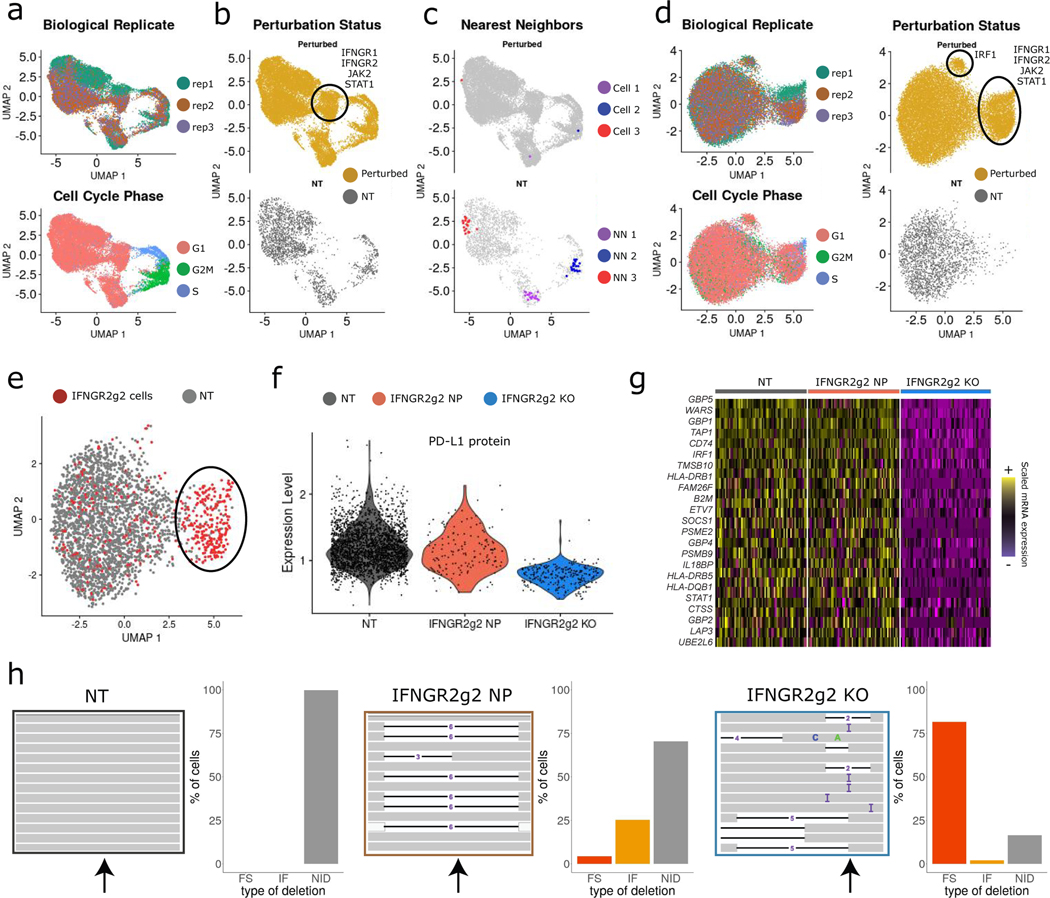
Figure 2. Calculating perturbation signature removes confounding variation.
a) UMAP visualization of the ECCITE-seq dataset based on cellular transcriptomes. Cells are colored by transduction replicate and cell cycle state. b) Same as in (a). Cells are split and colored by perturbation status (NT; non-targeted). Circle denotes a perturbation-specific cluster. c) Same as in (b). Top: example of three distinct cells expressing an IRF1 gRNA (red, blue, purple). Bottom: their 20 nearest NT neighbors (NN). Grey dots: all other cells. d) UMAP visualization based on perturbation signatures. Ovals denote perturbation-specific clusters. e) UMAP visualization showing IFNGR2g2 and NT cells. Oval denotes a group of putative IFNGR2g2 KO cells that cluster separately, but a subset of targeted cells appears to be non-perturbed (NP). f) Violin plot showing PD-L1 protein expression in NT, NP, and KO cells. IFNGR2g2 KO cells exhibit low PD-L1 protein levels while IFNGR2g2 NP and NT cells express PD-L1 at identical levels. g) Single-cell mRNA heatmap showing the IFNγ pathway-related gene expression in NT, NP, and KO cells. Gene expression is scaled (z-scored) across all single cells. For visualization purposes we downsampled our dataset to include 150 cells from each class. h) Interactive Genome Viewer (IGV) screenshot of a representative sample of reads mapping at theIFNGR2 gene locus (chr21: 34787276–34787299) targeted by IFNGR2g2 gRNA. CRISPR-induced INDELs are seen as black lines. Arrow indicates cut site. Barplote showing the % of IFNGR2 reads with no INDELS (NID), in-frame (IF) and frameshift (FS) mutations across NT (n=2,386), IFNGR2g2 NP and KO cells (n=278).