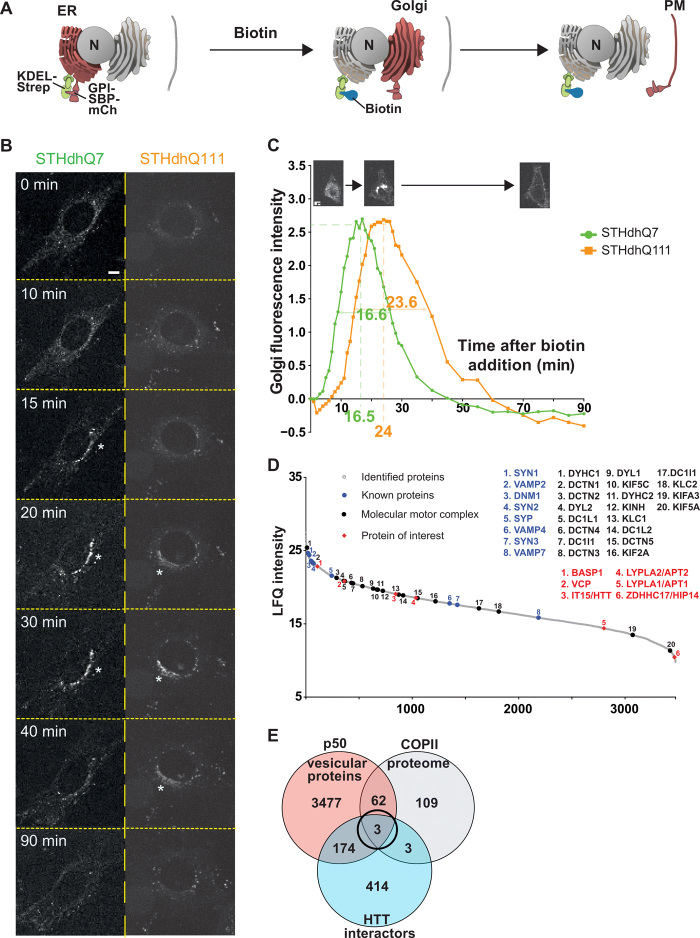
Fig. 1. Screening for modifiers of intracellular trafficking in Huntington disease.
(A) Schematic of the RUSH system KDEL-GPI-mCherry. GPI is fused with SBP and a fluorescent tag m-Cherry (mCh) (Reporter, red). The reporter is retained in the ER thanks to the Hook part (KDEL fused to streptavidin, green). The addition of biotin (blue) releases the reporter and allows trafficking of GPI-SBP-mCherry through the secretory pathway (Golgi and plasma membrane). (B) Localization of GPI-KDEL-mCherry at different time points after addition of biotin to STHdhQ7 and STHdhQ111 cells (scale bar, 10 μm). (C) Quantification of Golgi fluorescence in STHdhQ7 cells (green) and STHdhQ111 cells (orange). The green and orange numbers beneath the x axis and within the curves represent, respectively, the peak of Golgi fluorescence and the time to cross the Golgi in STHdhQ7 (N = 46) and STHdhQ111 cells (N = 54). (D) Proteins identified in the motile vesicle protein fraction were ranked by intensity and plotted according to their relative abundance (gray dots). Molecular motors and associated proteins (back dots) as well as previously identified vesicular proteins (blue dots) are among the most abundant proteins. HTT, HTT-interacting proteins present in the COPII proteome [brain abundant membrane attached signal protein 1 (BASP1), valosin-containing protein (VCP), and ZDHHC17], and APTs were identified as vesicular residents (red diamonds). (E) Venn diagram showing the overlap of proteomic (orange) and in silico screening of the HTT interactome (blue) and the COPII secretory pathway (gray) that identified three vesicular residents.