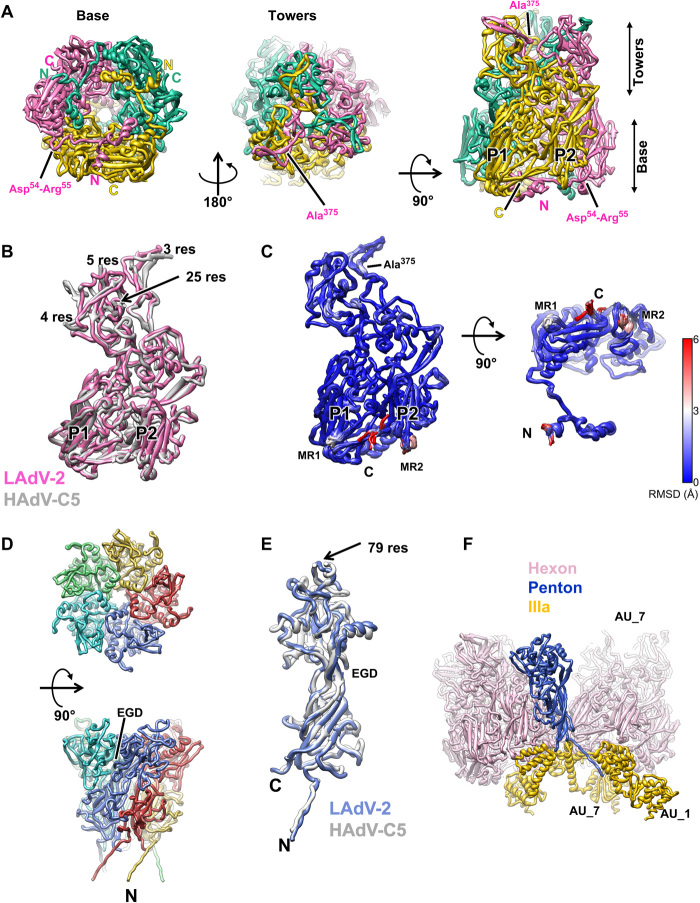
Fig. 2. LAdV-2 main capsid proteins: hexon and penton.
(A) Hexon trimer as seen from inside (left) or outside (center) the capsid and in a side view (right). The capsomer base and towers are indicated, as well as the N terminus (N) and C terminus (C) of each monomer. The two β barrels forming the double jelly roll are labeled P1 and P2. (B) Superposition of the LAdV-2 and HAdV-C5 (PDB ID 6B1T) hexon monomers. The length in residues of the HAdV-C5 untraced loops is indicated. (C) Superposition of the 12 hexon monomers in the LAdV-2 AU, colored by RMSD according to the scale at the right-hand side. MR1 and MR2: mobile regions. (D) Penton base pentamer as seen from outside the capsid (top) and in a side view (bottom). The N terminus of one monomer and the location of the EGD sequence are indicated. (E) Superposition of the LAdV-2 and HAdV-C5 penton base monomers. The length in residues (res) of the HAdV-C5 penton RGD loop is indicated. (F) Interactions of one penton base monomer with other proteins. AU_1 and AU_7 indicate molecules in neighboring AUs labeled as in fig. S3.