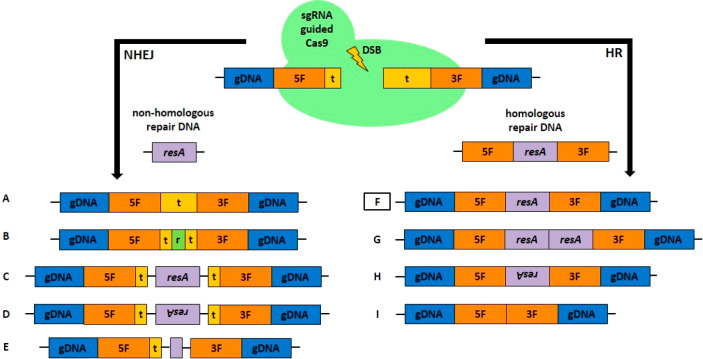
Figure 1.
Schematic overview of single guide RNA (sgRNA), guided Cas9 introduced DNA double-strand breaks (DSB), and possible DNA repair pathways in eukaryotes. Repair mechanisms include nonhomologous end joining (NHEJ) and homologous recombination (HR), the possible outcomes of which depend on the provision of DNA repair and the respective active repair mechanism. In the uppermost part, the selected exemplary nuclear (gDNA) target sequence (t) and 5′ and 3′ flanking regions (5F, 3F) are depicted. Using sgRNA-guided Cas9, a DSB can be introduced in the target region, but the process is not entirely predictable. As long as a nonhomologous repair template confers antibiotic resistance (resA cassette), an active NHEJ pathway can lead to several results: (A) target sequence repair with error-prone sequence modifications, (B) insertion of random (r) sequences, (C) insertion of DNA clockwise, and anticlockwise in (D), and (E) insertion of truncated repair DNA. As a consequence, there are likely to be remnants of the chosen target sequence. In contrast, the provision of homologous repair DNA containing the flanking regions can lead to different results in an active HR pathway: the most desirable outcome is shown in (F), that is, the sequence-specific replacement of the target sequence with resA. Other options are multiple insertions of resA (G), inverted insertions (H) and deletions (I).