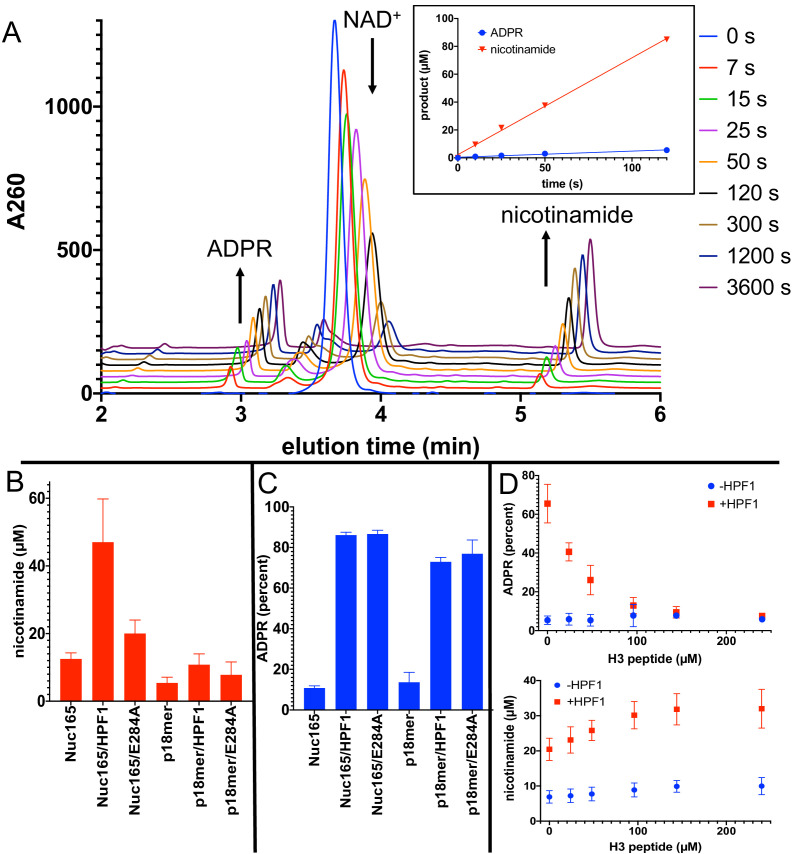
Figure 5. Addition of Histone PARylation Factor 1 (HPF1) converts poly(ADP-ribose) polymerase 1 (PARP1) predominantly into an NAD+ hydrolyase.
(A) Representative HPLC traces from a reaction of PARP1 using p18 as an activator that simultaneously monitors depletion of NAD+ (200 µM initial) and formation of both ADP-ribose (ADPR) and nicotinamide. Each successive time point trace is offset in both the x- and y-axis to allow for better visualization. The inset shows that the assay is linear with respect to formation of nicotinamide and ADPR for at least 90 s. (B) Comparison of activity of PARP1 (100 nM) as measured by formation of nicotinamide using either Nuc165 (300 nM) or p18mer (100 nM) as activators in the presence or absence of HPF1 (2 µM, wild-type vs. E284A mutant) after 1 min of reaction time. (C) Comparison of percent of turnover of PARP1 that leads to free ADPR under the same conditions as in (B). Error bars in (C) and (D) are derived from three experiments, each performed using four replicates. (D) The hydrolase activity of PARP1 (100 nM) is suppressed (top panel) with a modest increase in overall activity as measured by nicotinamide formation (bottom panel) by high concentrations of H3 histone tail peptide (varied as indicated), but only in the presence of HPF1 (2 µM). Reactions were performed at 200 µM NAD+ for 1 min, and the data and standard deviations shown are derived from four separate experiments.