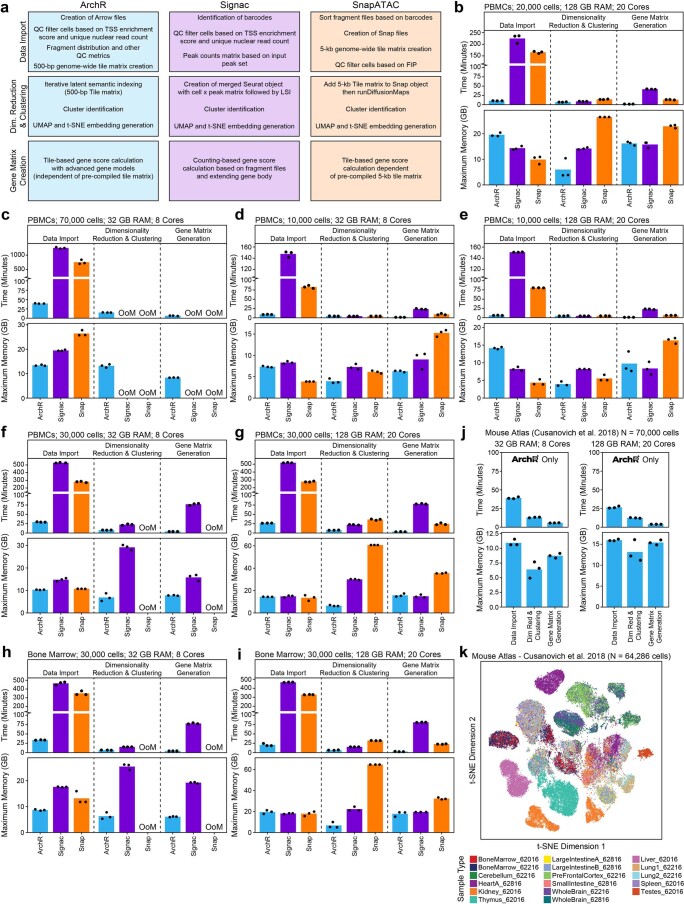
Extended Data Fig. 2. Benchmarking comparisons of runtime and memory usage for ArchR, Signac, and SnapATAC.
a, Schematic describing the individual benchmarking steps compared across ArchR (blue), Signac (purple), and SnapATAC (orange) for (1) Data Import, (2) Dimensionality Reduction and Clustering, and (3) Gene Score Matrix Creation. b-i, Comparison of ArchR, Signac, and SnapATAC for run time and peak memory usage for the analysis of (b) ~20,000 cells from the PBMCs dataset using 128 GB of RAM and 20 cores (plot corresponds to Fig. 1b), (c) ~70,000 cells from the PBMCs dataset using 32 GB of RAM and 8 cores (plot corresponds to Fig. 1c), (d-e) ~10,000 cells from the PBMCs dataset using (d) 32 GB of RAM and 8 cores or (e) 128 GB of RAM and 20 cores, (f-g) ~30,000 cells from the PBMCs dataset using (f) 32 GB of RAM and 8 cores or (g) 128 GB of RAM and 20 cores, and (h-i) ~30,000 cells from the bone marrow dataset using (h) 32 GB of RAM and 8 cores or (i) 128 GB of RAM and 20 cores. Dots represent individual replicates of benchmarking analysis (N = 3). OoM corresponds to out of memory. j, Benchmarks from ArchR for run time and peak memory usage for the analysis of ~70,000 cells from the sci-ATAC-seq mouse atlas dataset (N = 13 tissues) for (left) 32 GB of RAM with 8 cores and (right) 128 GB of RAM with 20 cores. Dots represent individual replicates of benchmarking analysis. k, t-SNE of mouse atlas scATAC-seq data (N = 64,286 cells) colored by individual samples.