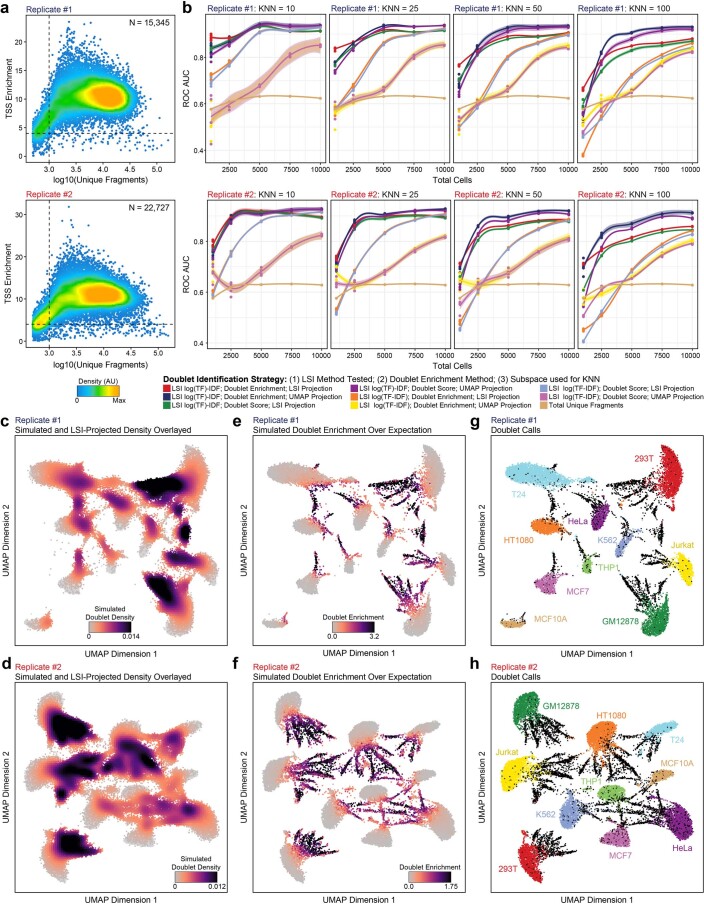
Extended Data Fig. 3. Optimization of doublet identification using admixtures of cell lines.
a, QC filtering plots from ArchR for (top) replicate 1 and (bottom) replicate 2 from the cell line mixing dataset showing the TSS enrichment score vs unique nuclear fragments per cell. Dot color represents the density in arbitrary units of points in the plot. b, Accuracy of various doublet prediction methods for (top) replicate 1 and (bottom) replicate 2 from the cell line mixing dataset, measured by the area under the curve (AUC) of the receiver operating characteristic (ROC), across different in silico cell loadings. Accuracy is determined with respect to genotype-based identification of doublets using demuxlet. Above each plot, ‘KNN’ represents the number of cells nearby each projected synthetic doublet to record when calculating doublet enrichment scores. The distance for KNN recording is determined in the LSI subspace for LSI projection and in the UMAP embedding for UMAP projection parameters. The smooth line represents a LOESS fit (shading represents 95% confidence interval). c-h, UMAP of scATAC-seq data showing the (c-d) simulated doublet density, (e-f) simulated doublet enrichment, or (g-h) cell line identity based on genotyping information and demuxlet for (c,e,g) replicate 1 (N = 15,345 cells) and (d,f,h) replicate 2 (N = 22,727 cells) of the cell line mixing dataset.