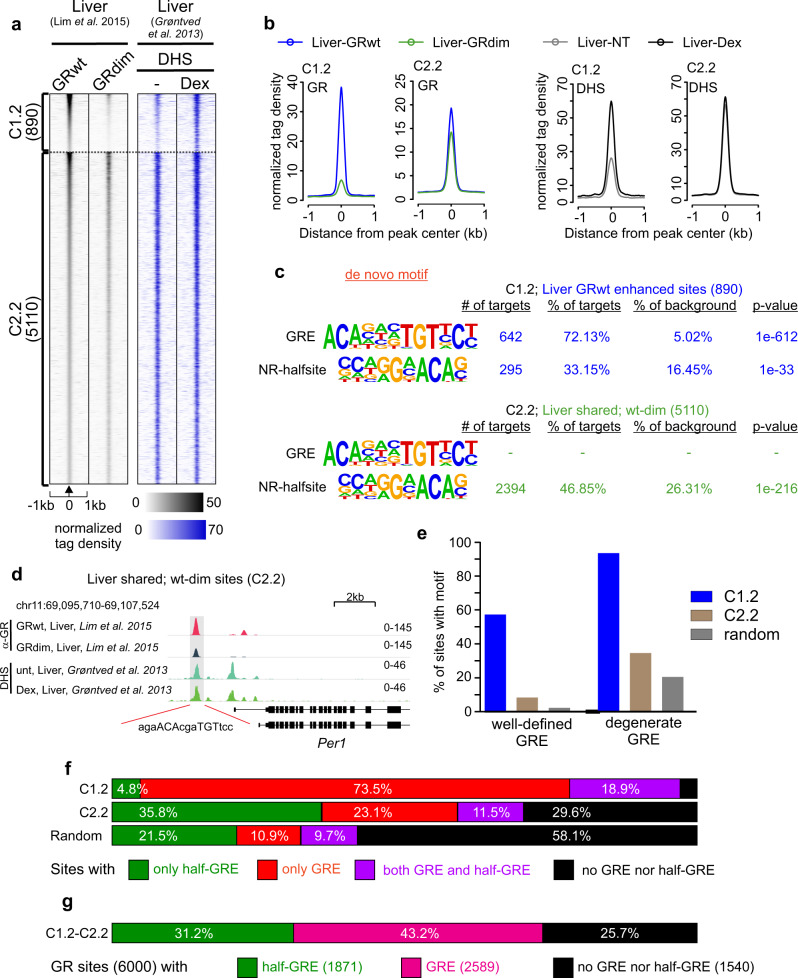
Fig. 5. Liver GR chromatin binding and de novo motif analyses.
a Comparison of GRwt and GRdim ChIP-seq data (black) in mouse liver reveals two clusters, C1.2 (GRwt specific) and C2.2 (GRwt/GRdim shared). DNase I hypersensitive sites (DHS) (blue) from mouse liver also shown. Each heat map represents ±1 kb around the center of the GR peak. Binding intensity scale noted below on a linear scale and treatment noted above for DHS. Heat maps sorted based on GRwt binding intensity and normalized to 10 million reads and to local tag density. b Aggregate plots of GRwt (blue) and GRdim (green) binding for each cluster and the corresponding DHS (GRwt-black, GRdim-gray). All aggregate plots normalized to 10 million reads, and further to local tag density (tags per site per bp). c De novo motif analysis shows the percentage of sites with a motif and the calculated background percentage from 50 K random sites (with matched GC% content), p-value of the motif enrichment. The PWM logo of detected motif shown on left. d Genome browser track of Per1 locus with perfect GRE (gray highlight) and GR binding sites in each cell line. Tracks are normalized to total of 10 million reads. e The percentage of sites within each cluster that contain each pre-defined motif (see Fig. 4b) as well as the percentage at 2000 randomly chosen DNase-seq peaks from the mouse liver. f The percentage of GR peaks with only a full GRE (red color), only a half-GRE (green color), both a full GRE and half-GRE (purple color), or no GRE nor half-GRE (black color). The degenerate full GRE and half-GRE in Fig. 3a were used to determine the percentages. g The percentage of all GR peaks (6000) that contain half-GRE (green), full GRE (magenta) or no half nor full GRE (black). Source data are provided as a Source Data file.