Abstract
The Serine Protease Inhibitor (serpin) protein has been suggested to play a key role in the interaction of bifidobacteria with the host. By inhibiting intestinal serine proteases, it might allow bifidobacteria to reside in specific gut niches. In inflammatory diseases where serine proteases contribute to the innate defense mechanism of the host, serpin may dampen the damaging effects of inflammation. In view of the beneficial roles of this protein, it is important to understand how its production is regulated. Here we demonstrate that Bifidobacterium longum NCC 2705 serpin production is tightly regulated by carbohydrates. Galactose and fructose increase the production of this protein while glucose prevents it, suggesting the involvement of catabolite repression. We identified that di- and oligosaccharides containing galactose (GOS) and fructose (FOS) moieties, including the human milk oligosaccharide Lacto-N-tetraose (LNT), are able to activate serpin production. Moreover, we show that the carbohydrate mediated regulation is conserved within B. longum subsp. longum strains but not in other bifidobacterial taxons harboring the serpin coding gene, highlighting that the serpin regulation circuits are not only species- but also subspecies- specific. Our work demonstrates that environmental conditions can modulate expression of an important effector molecule of B. longum, having potential important implications for probiotic manufacturing and supporting the postulated role of serpin in the ability of bifidobacteria to colonize the intestinal tract.
Subject terms: Bacterial physiology, Microbiology
Introduction
Bifidobacterium longum subsp. longum is a Gram-positive, high G+C content, anaerobic bacterium that is a member of the phylum Actinobacteria. A variety of bifidobacteria species are natural inhabitants of the human gastro-intestinal tract (GIT). In particular, they are dominantly colonizing the infant gut. Bifidobacteria are believed to be acquired by vertical transmission from the mother and their persistence in the infant gut is associated with their saccharolytic activity toward the abundant host and diet-derived glycans present in the infant gut1. Human Milk Oligosaccharides (HMOs) are complex carbohydrates found in human milk where they are the third most-abundant solid component2. HMOs, as well as lactose, galacto-3,4 and fructo-oligosaccharides5 in infant formula have been shown to drive Bifidobacterium enrichment in the infant gut. In addition, bifidobacteria remain present in the human GIT throughout life and various prebiotic dietary supplements (including galacto- and fructo- oligosaccharides) also enrich the bifidobacterial populations in adulthood. According to recent estimates and depending on the geographical location, B. longum subsp. longum can make up to 20 % of the Bifidobacterium community in the intestine, which can constitute up to 4% of the overall microbiota in adults1. Strains belonging to the Bifidobacterium genus, including those belonging to B. longum subsp. longum, are widely used as probiotics and their beneficial effects on specific intestinal and extra-intestinal pathologies have been documented6.
The genome of the strain B. longum subsp. longum NCC 2705 (hereafter B. longum NCC 2705), isolated from infant feces, highlighted its particular adaptation to saccharolytic metabolism relevant in the human gut environment, as illustrated by a large repertoire of genes encoding carbohydrate degrading enzymes and import systems7,8. The strain also contains different systems enabling it to resist and evolve in the competitive GIT environment. For example, the genome encodes a bile salt hydrolase and the corresponding efflux system conferring bile acid resistance9, as well as genes encoding fimbriae that have been shown to drive its in vitro adhesion to mucins10. This strain is also able to limit growth of the pathogenic E. coli O157 in vivo through the production of acetate, which depends on the gene encoding the sugar ABC transporter solute-binding protein (BL0033)11. Moreover, B. longum NCC 2705 produces a serine protease inhibitor (serpin) encoded by the BL0108 gene, which forms covalent products with pancreatic and neutrophil elastases thereby inhibiting their function12. This serpin is proposed to play an important role in the colonization of bifidobacteria by protecting them against host-derived proteases and providing them with a survival advantage in the competitive intestinal environment12,13. The serpin’s capacity to inhibit the Human Neutrophil Elastase12 may also be involved in the immunomodulatory capacities of the strain14 as elastase is released by activated neutrophils at the sites of intestinal inflammation15. In line with this role in dampening innate immunity, serpin was demonstrated to play a key role in the anti-inflammatory effect of B. longum NCC 2705 in a mouse model of gluten sensitivity16. Recently, the serpin of the NCC 2705 strain was reported to prevent enteric nerve activation in vitro, which suggest a potential role of this protein in pain reduction in Irritable Bowel Syndrome patients17.
In view of the hypothesized colonization and bioactive roles of serpin in bifidobacteria, it is important to understand how its production is regulated. Serpin encoding genes have been identified in a limited number of bifidobacterial species, including B. breve, B. longum subsp. longum, B. longum subsp. infantis, B. longum subsp. suis and B. dentium13. However, querying the genomes of a range of recently described bifidobacterial species18 identified several additional serpin-encoding genes, suggesting that the function is more widely spread among the members of this genus (Table S1). A previous study failed to detect serpin production by western blot in MRSc grown B. longum NCC 2705 and serpin was thus considered to be induced in vivo by unknown factors (F. Arigoni, personal communications). Transcriptional regulation studies of the B. breve serpin-encoding gene showed that it involves a protease inducible two-component system located next to the serpin encoding operon19. This system is absent from B. longum subsp longum strains and is partially present in strains of B. longum subsp. infantis. In all studied species, it was shown that the serpin encoding gene is flanked by genes encoding a Lac-I regulator and a membrane-associated protein of unknown function, although the latter gene is not universally conserved13. The observed variations in the gene syntenies around the serpin gene suggest that its regulation may not only differ in different species, but may even be subspecies specific13. We have here revealed that different carbohydrates control production of serpin, a protein playing a key role in the bioactivity of B. longum subsp. longum.
Results
Development of a sandwich ELISA enabling serpin quantification
Polyclonal antibodies obtained by immunizing rabbits with the previously purified B. longum NCC 2705 serpin recombinant protein12 were used to develop a sandwich ELISA (Figure S1). This method enabled the accurate quantification of the serpin protein in subcellular fractions and crude lysates of B. longum NCC 2705, with a limit of quantification (LoQ) of 4 pg/ml of serpin. Moreover, it allowed the detection and quantification of the serpin from B. breve ATCC 15700, a protein with only 93% sequence identity compared to the B. longum subsp. longum serpin13 (Figure 1).
Figure 1.
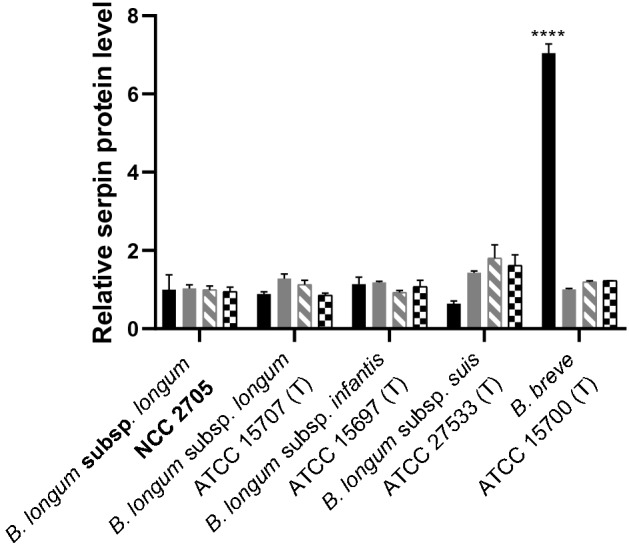
Effect of different proteases (papain [black bars], trypsin [grey bars], chymotrypsin [dashed gray bars] and pig pancreatic elastase [PPE, squared black bars]) on the serpin production levels in bifidobacteria harboring the serpin encoding gene, after 16 h of growth. The studied strains are B. longum subsp. longum NCC 2705, B. longum subsp longum ATCC 15707 (type strain [T]), B. longum subsp. infantis ATCC 15697 (T), B. longum subsp. suis ATCC 27533 (T) and B. breve ATCC 15700 (T). Serpin levels are expressed relative to the control level (including standard deviations) measured in MRS without addition of proteases. Statistical difference relative to the control cells are indicated; ****p < 0.0001.
Localization of Serpin in B. longum NCC 2705
Examination of the B. longum NCC 2705 BL0108 serpin protein sequence using Interpro-scan20 predicted that the protein contains a N-terminal transmembrane domain12. Analysis by the Signal P21 package did not predict a signal peptidase cleavage site (Figure S2), suggesting that the protein would be N-terminally anchored by a transmembrane domain. We investigated serpin localization by the comparative analysis of serpin abundance in cell-associated (cytoplasmic, cell-wall) and the spent culture supernatant (secreted protein) fractions (Figure 2). This analysis revealed that both in the wildtype strain B. longum NCC 2705 and in its pMDY25-harboring serpin overexpressing derivative16, the serpin protein remains associated with the cells and is not released in the environment which in part confirms the predicted localization. The level of serpin produced by the recombinant strain is drastically (> 10,000-fold) higher than the level found in its wildtype counterpart. Importantly, no serpin could be detected in cell-associated or supernatant fractions from strain B. longum subsp. longum NCC 9035, a genetically engineered serpin null-mutant of NCC 270516 (Figure 2). The minimal background reactivity observed in the cell-wall associated fraction of the NCC 9035, was assumed to be due to minor cross-reactivity of the polyclonal antibody used.
Figure 2.
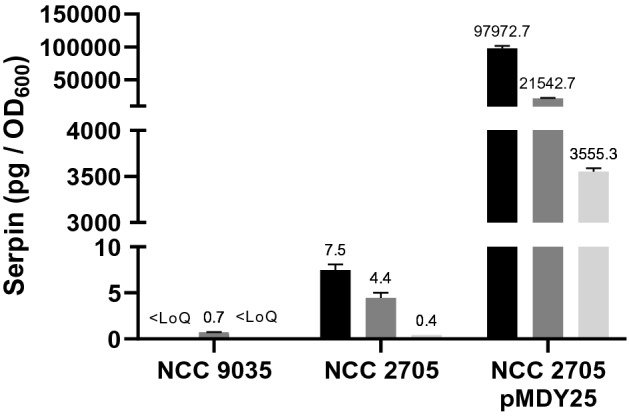
Serpin concentration and localization in extracts of B. longum NCC 2705, its serpin knock-out counterpart (NCC 9035) and the constitutively producing (NCC 2705 harboring pMDY25) recombinant strain. Means and standard deviations are depicted. Black bars represent cytoplasmic fractions, dark grey bars represent cell-wall associated fractions and light grey represent supernatant fractions (i.e. excreted protein). Serpin amounts are reported in picograms (pg) per mg of total protein.
Evaluation of extracellular proteases as inducing factor for serpin production
B. longum subsp. longum NCC 2705, B. longum subsp. longum ATCC 15707 (type strain [T]), B. longum subsp. infantis ATCC 15697 (T), B. longum subsp. suis ATCC 27533 (T) and B. breve ATCC 15700 (T), all belonging to different non-pathogenic species that encode a serpin orthologue closely related to Bl0108 protein (with % of identity of 99.8, 94.9, 92.7 and 93.3 respectively), were grown in presence of different proteases (papain, trypsin, chymotrypsin and pig pancreatic elastase) previously shown to modulate serpin mRNA levels13,19. Cell-exposure to papain induced a 7-fold increase in serpin production in B. breve ATCC 15700, which is in agreement with the previously reported increased transcription of the serpin gene in this strain19. Conversely, none of the studied proteases led to a significant increase in the levels of cell associated serpin in B. longum subsp. longum, B. longum subsp infantis and B. longum subsp. suis strains, indicating that serpin production is differentially regulated among these bifidobacterial species (Figure 1).
Galactose and fructose driven induction of B. longum NCC 2705 serpin is repressed by glucose
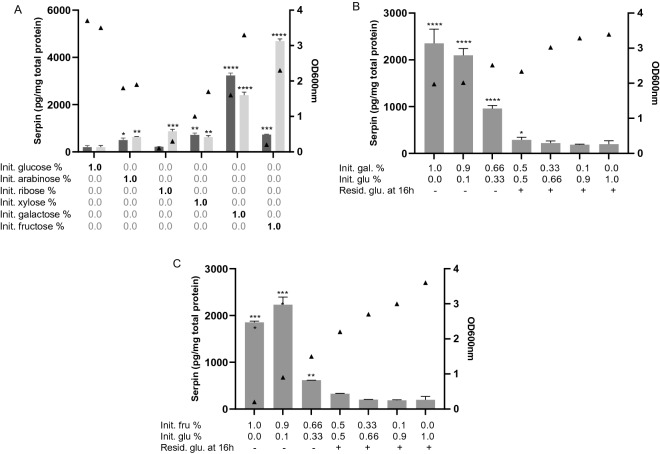
To test the capacity of different carbohydrates to induce serpin production in B. longum NCC 2705 the strain was grown on carbohydrate-free MRSc medium supplemented with 1% (w/v) of different monosaccharides previously shown to support its growth, i.e., glucose, arabinose, ribose, xylose, galactose and fructose8. Growth on the majority of the sugars tested led to a significant increase of serpin levels compared to those measured after growth on glucose (Figure 3A). While only a modest increase in serpin production was observed when the strain was grown on the pentoses arabinose, ribose and xylose, growth on hexoses led to strongly increased levels of serpin, i.e., 16- and 24-fold increase after growth on galactose and fructose, respectively. In these experiments all media were inoculated using a glucose grown preculture which explains the delayed growth observed in some of the conditions. The relatively poor overnight growth observed for B. longum NCC 2705 on fructose-media is reflecting the slow-adaptation of the strain to the utilization of this sugar. This was further confirmed by extending the incubation on fructose-media to 48 hours and reaching high OD600 values (Figure 3A). Nevertheless, despite the relatively poor growth on fructose media in the initial overnight culture, these growth conditions led to a strongly increased level of serpin (Figure 3A,C).
Figure 3.
Serpin concentration measured in extracts of B. longum NCC 2705 grown for 24 (dark grey) and 48 h (light grey) on different monosaccharides (A). Serpin concentration measured in B. longum NCC 2705 grown for 16 h on different concentrations of glucose and galactose (B) and glucose and fructose (C). Bars represent serpin quantity normalized by total protein content, means and standard deviations are depicted. Initial carbohydrate concentrations are depicted. Presence of residual glucose in the culture supernatant after 16 h of incubation is indicated (+ /−) (B,C). Triangles represent growth at the end of the incubation, measured by optical density at 600 nm. P-values represent statistical difference to the glucose control group. ****p < 0.0001; ***p < 0.001; **p < 0.01; *p < 0.05.
To assess the potential inhibitory effect of glucose, serpin production was next measured after growth of B. longum NCC 2705 in media containing different ratios of glucose:galactose (Figure 3B) and glucose:fructose (Figure 3C). In both set of experiments, optical density at 600 nm (OD600) and residual glucose levels in the supernatant were measured after 16h of incubation. We observed that the presence of glucose in the medium prevented the galactose- and fructose-mediated induction, which could be indicative of glucose-mediated catabolite repression. Once the glucose was depleted from the medium (Figure 3B,C), the incubation in galactose and fructose media led to induction of serpin production. Notably, the induction of serpin production in fructose media did not appear to depend on growth on this substrate (Figure 3C).
Galactose induction mechanism is conserved in B. longum subsp. longum strains
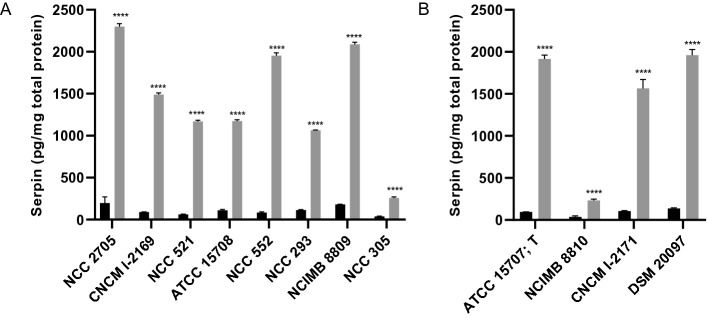
Different B. longum subsp. longum strains (Table S2) were selected to represent the phylogenetic diversity of this subspecies, and these strains were grown on galactose as a sole carbon source for 16h. In these experiments galactose containing media were inoculated with cultures grown in glucose medium. Several strains displayed efficient growth on galactose under these conditions (NCC 2705, CNCM I-2169, NCC 521, ATCC 15708, NCC 552, NCC 293, NCIMB 8809, NCC 305), and in all these cultures a significantly increased level of serpin was detected as compared to their glucose-grown counterparts (Figure 4A). However, galactose did not support growth of all strains within 16h of incubation (ATCC 15707, NCIMB 8810, CNCM I-2171 and DSM 20097), which could reflect strain-specific inability to utilize galactose or a longer lag phase when switching from glucose to galactose. For these strains, a small amount of glucose (0.2%) was added to the galactose-containing (0.8%) medium to allow the production of biomass during the first 16h of incubation, while also including a prolonged period of incubation in galactose containing medium when glucose was depleted. Under these conditions, incubation in galactose containing medium led to increased serpin production (Figure 4B). These results demonstrate that galactose is consistently inducing serpin production in B. longum subsp. longum strains even in strains that are unable to grow efficiently on this monosaccharide as a sole carbon source.
Figure 4.
Serpin protein concentration in a set of B. longum subsp. longum strains grown for 16 h on MRSc medium supplemented with 1% glucose (black bars) or 1% galactose (grey bars). Bars represent means and standard deviations are shown. Panel A depicts B. longum subsp. longum strains able to rapidly grow on galactose as a sole carbon source). Panel B displays B. longum subp. longum strains not able to rapidly switch to galactose as a sole carbon source. These strains were grown on a medium supplemented with 1% glucose (black bars) or with a mixture of 0.2% glucose and 0.8% galactose (grey bars). P-values represent statistical difference to the glucose control groups. ****p < 0.0001.
Similar experiments were performed to evaluate the conservation of the observed serpin regulation in other Bifidobacterium species, and/or subspecies of B. longum, revealing that galactose-induced serpin production is conserved in B. longum subsp. suis ATCC 27533 (T) (10.8 fold compared to glucose grown culture) but not in B. longum subsp. infantis ATCC 15697 (T) or B. breve ATCC 15700 (T) (Figure S3). This demonstrates that differential serpin regulation is not only observed when comparing different Bifidobacterium species, but also among different B. longum subspecies.
Fructose and galactose containing di- and oligosaccharides induce serpin production
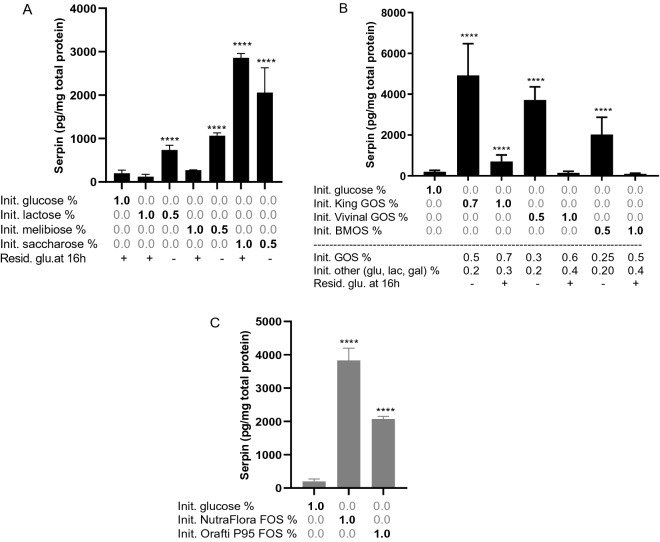
B. longum NCC 2705 was grown to stationary phase on MRSc medium supplemented with different galactose or fructose containing disaccharides (lactose, melibiose and saccharose), galacto- or fructo-oligosaccharides (GOS and FOS), or human milk oligosaccharides (HMOs). Serpin production was quantified in crude cell extracts derived from the resulting cultures.
Growth on lactose (Gal-β[1→4]-Glc) and melibiose (Gal-α[1→6]-Glc) induced serpin production (respectively 3.7 and 5.2 fold compared to growth on glucose) when supplemented at a concentration of 0.5% (w/v) but not at 1.0 % (w/v). In the latter condition, glucose generated by the hydrolysis of the disaccharides was not depleted at harvesting (16h, Figure 5A). In contrast, saccharose (Glc-α[1→2]-Fru), a fructose-containing disaccharide led to increased levels of serpin irrespective of the concentration used (0.5% or 1.0% supplementation) (Figure 5A).
Figure 5.
Serpin concentration in extracts of B. longum NCC 2705 grown to the stationary phase (16 h) on MRSc medium, supplemented with the di-saccharides galactose, melibiose and saccharose (panel A), different galacto-oligosaccharides (GOS) (panel B), or different fructo-oligosaccarides (FOS) (panel C). Bars represent means and standard deviations are depicted. Initial carbohydrate concentrations as well as the presence of residual glucose at harvesting are indicated (A,B). P-values represent statistical difference to the glucose control groups. ****p < 0.0001.
Growth on different GOS preparations led to a maximal 25-fold induction of serpin in B. longum NCC 2705 (Figure 5B). The GOS preparations contain relatively high level of glucose and lactose (King GOS, 28%; Vivinal GOS, 40%; BMOS, 38%). Consequently, also for these substrates the serpin inducing effect was lost at higher supplementation concentrations, correlating with the presence of residual glucose at the time of harvest (16h) (Figure 5B). The FOS preparations tested are purer than GOS and do not contain glucose, explaining why these substrates consistently induced serpin production in B. longum NCC 2705 irrespective of the supplementation concentration (Figure 5C).
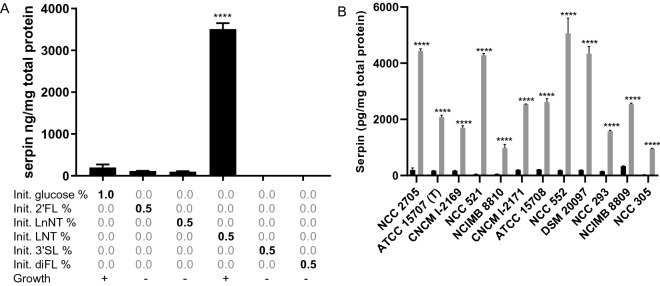
Finally, B. longum NCC 2705 was grown in media containing 0.5% of different HMOs (2’FL, LnNT, LNT, 3’SL and diFL) as sole carbon source. The strain could only grow on Lacto-N-tetraose (LNT), which also led to a 18-fold induction of serpin production as compared to glucose (Figure 6A). LNT was subsequently shown to consistently support the growth and induce serpin production in the different strains representing the phylogenetic diversity of the B. longum subsp. longum subspecies (Figure 6B).
Figure 6.
Serpin concentrations in extracts of B. longum NCC 2705 grown to the stationary phase (16 h) on MRSc medium, supplemented with the human milk oligo-saccharides (2′-O-Fucosyllactose [2′FL], Lacto-N-neotetraose [LNnT], Lacto-N-tetraose [LNT], 3′-O-Sialyllactose [3′SL], Difucoxyllactose [DiFL]) (panel A). Different B. longum subsp. longum strains grown to stationary phase(16 h) on MRSc supplemented with 1% glucose (black bars) or 0.5% LNT (grey bars) (panel B). Bars represent means and standard deviations are depicted. P-values represent statistical difference to the glucose control groups. ****p < 0.0001.
Taken together these results establish that growth of B. longum NCC 2705 (and several other strains of the same subspecies) on galactose and fructose containing di- and oligosaccharides consistently led to increased serpin production. This induction was observed only when the glucose moieties of the galactose containing substrates were completely depleted, which was not the case for the fructose containing carbohydrates.
AraQ involvement in serpin regulation?
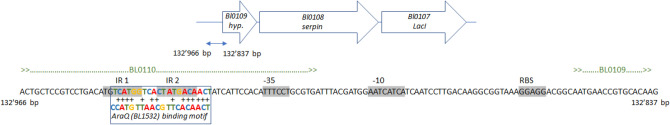
A MAST search using all transcriptional factors predicted to be implicated in the carbohydrate metabolism of B. longum NCC 2705 enabled to identify three LacI-type regulator binding sites in the previously predicted promoter region upstream of the Bl0109 gene13. Two binding sites resembling Bl0185 (BL0187) and GosR (BL0258) motifs were located far away from the start of the Bl0109 gene (respectively 692 and 1107 bp). In contrast, a motif resembling the predicted AraQ (BL1532) binding site was found at a reasonable distance from the serpin operon (94 bp upstream from the Bl0109 start-codon) (Table 1). The resemblance of the potential AraQ binding site is modest when compared to the original BL1532 AraQ binding site but its close localization to the predicted -35 promoter-element supports a role in serpin operon regulation (Figure 7).
Table 1.
List of transcription factor binding sites predicted by MAST upfront the promoter region of the BL0109 gene.
| Regulon name | Regulon type | Original sequence (from RegPrecise) | Target sequence upfront of serpin operon | N° of mismatches | Distance from Bl0109 start (bp) | p-value obtained from MAST |
|---|---|---|---|---|---|---|
| AraQ (BL1532) | LacI | CCATGTTAACGTTCACAACT | TCATGGTCACTATGACAACT | 6 | − 94 | 3.5E−05 |
| BL0185 (BL0187) | LacI | ATATTGCATCGATGTAAATA | CTATTGCATCGATGCTGATT | 5 | − 692 | 6.9E−06 |
| GosR (BL0258) | LacI | TTGGTCAACCGGTGTATCAA | CTGGTCAAGCGTGGTATTAA | 5 | − 1107 | 4.1E−06 |
Figure 7.
Genetic setup of the serpin operon promoter as previously described by Turroni et al. Inverted repeats (IR 1 and IR 2), -35 and -10 hexamers as well as Ribosome Binding Site (RBS) are depicted in grey. The motif identified using MAST and resembling the previously predicted AraQ motif ahead of the BL1532 gene is depicted with bold colors.
Discussion
In this work, we have developed a sensitive ELISA assay which enabled to quantify levels of serpin present in different bifidobacterial species. Using this assay, we could confirm the high levels of serpin produced by the serpin overexpressing pMDY25-harboring derivative of B. longum NCC 270516 as well as the lack of serpin detection in the serpin-null mutant B. longum NCC 9035 previously used to decipher the role of serpin in a mouse model of celiac disease16. Serpin localization experiments using B. longum NCC 2705 showed that the protein is retained in the cellular biomass and not secreted to the medium, which confirmed the predicted N-terminal anchoring of the protein in the cell membrane in combination with the lack of a recognizable signal peptidase cleavage site. However, serpin release in the medium was more prominently detected in the culture of the serpin-overproducing B. longum NCC 2705 that harbors pMDY25, which is probably reflecting hampered protein biogenesis as a consequence of the very high serpin production in this strain. Nevertheless, the results establish that to asses regulation of serpin production levels by culture conditions, the protein is best quantified by evaluating its specific-level in the cellular biomass and can ignore the very minor amounts that are released into the medium.
Serpin encoding genes have been found to be conserved among a subset of bifidobacterial species13. Previous studies have reported that its transcription in B. breve is induced by treatment of the cells with proteases, in particular papain, involving a two-component regulatory system, which is not encoded in B. longum13,19. The observation in B. breve is in apparent agreement with the proposed role of serpin in protection of the bacterial cell against the detrimental impact of environmental proteases. Our results confirmed that serpin production is induced by papain treatment in B. breve and established that a similar treatment with papain or other proteases failed to induce it in various B. longum subspecies (longum, infantis, and suis). Notably, we could establish that serpin production in B. longum subsp. longum and B. longum subsp. suis is controlled by the carbon source in the culture medium, identifying galactose and fructose as potent inducers. Furthermore, our results suggest that import of these carbohydrates in the cell is essential for their effect on serpin production, even though their metabolization appears not required (Figures 3C, 4B). None of the carbon sources tested was able to modulate serpin levels in B. longum subsp. infantis. This highlights that diverse mechanisms regulate production of serpin in different bifidobacterial species (e.g. B. breve versus B. longum), but also in different subspecies. The regulation of serpin production by carbon sources in specific B. longum subspecies does not have an obvious relation with the anti-protease function of serpin. This may suggest that an additional functional role of this protein remains to be deciphered.
The data we present suggest the involvement of catabolite repression in the control of serpin production. Growth of B. longum on the pentoses arabinose, ribose and xylose led to a modest but statistically significant increase of serpin production compared to glucose, which may be due to relieved glucose catabolite repression. This is in sharp contrast to the prominent serpin-inducing capacity of the hexoses galactose and fructose. Even though both pentoses and hexoses are metabolized through the so called “bifid-shunt”, their metabolism is markedly different; pentoses are entering the energy generating phosphoketolase-dependent pathway as glyceraldehyde-3-phosphate, while hexoses are converted by phosphoglucomutase to glucose-6-phosphate and fructose-6-phosphate22. Metabolic intermediates of the “bifid-shunt” may play an important role in the regulation of serpin and would deserve to be further studied in this context.
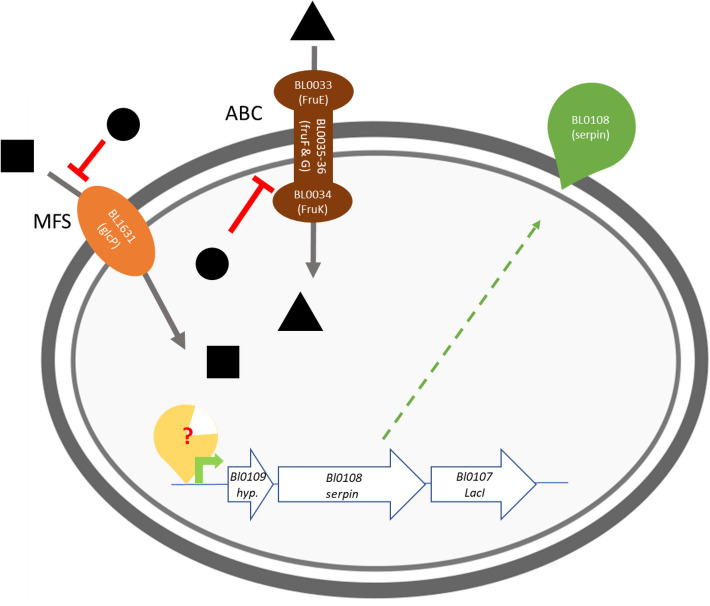
Although the precise molecular mechanisms involved in serpin regulation in B. longum NCC 2705 remains to be elucidated, we could demonstrate that galactose- and fructose-mediated serpin induction is prevented by presence of glucose in the environment. In B. longum NCC 2705, the glucose/mannose transporter protein (encoded by BL1631; glcP) is involved in galactose import8. This protein was demonstrated to have the highest specificity for glucose followed by mannose and galactose23, which could explain why glucose depletion is required to enable the subsequent import of galactose that once internalized could play a role in the intracellular activation mechanism of serpin production. In the same strain, an ATP-binding cassette transporter encoded by the operon Bl0033-0036 (fruEKFG) mediates fructose import. Previous studies have shown that the synthesis of the substrate binding protein (FruE) is strongly suppressed by the presence of glucose24, suggesting that the fru operon is suppressed by the presence of glucose in the medium. Thereby, the presence of glucose in the medium could inhibit fructose-import, and assuming that internalized fructose (analogous to internalized galactose) could play a role in in the intracellular serpin production-activating regulatory mechanism provides a rational explanation of glucose suppression of fructose-mediated activation of serpin production (Figure 8).
Figure 8.
Proposed schematic representation of serpin induction by galactose or fructose and its repression by glucose. Presence of glucose (black circles) inhibits transport of galactose (black squares), as it is known to have a higher affinity to the Major Facilitator Superfamily (MFS) transport protein glcP protein (BL1631). As well, presence of glucose in the cell has been previously shown to inhibit the production of the ATP-binding cassette transporters (ABC) encoded by the genes located in the operon BL0033-BL0036 and responsible of fructose (black triangles) import. The exact regulatory genetic mechanism enabling serpin production remains to be elucidated.
These results resemble typical manifestations of glucose mediated catabolite repression, which is well described in many bacteria25 and for which a few examples have been described in bifidobacteria23,26,27. In most Gram positive bacteria, catabolite repression is controlled by the complex formed by the histidine phosphocarrier protein (HPr) and catabolite control protein A (CcpA), a LacI-type regulator that can bind to specific DNA motifs (so-called cre-elements) and modify transcription of downstream genes28. At present, the closest relatives of the Bacillus subtilis CcpA in B. longum NCC 2705 are annotated as LacI-type regulators, and their homology with CcpA is quite low (24-32% identity). Bifidobacteria encode a large number of LacI-type regulators known to commonly regulate carbohydrate metabolism. For example, B. longum NCC 2705 harbors 16 genes encoding LacI-regulators according to the RegPrecise database29. Most of these LacI-type regulators are foreseen to act locally by regulating genes located in their vicinity. This is exemplified by the LacI encoding gene BL0107 that is genetically linked to the serpin encoding gene and is predicted to regulate the closely located sucrose utilization operon that encodes a sucrose permease (BL0106) and a sucrase (BL0105). Recently, two LacI-type transcriptional regulators, AraQ and MalR1, were demonstrated to control a large set of genes spread over the genome of B. breve UCC 2003 including genes involved in the”bifid-shunt” as well as transcription factors, and uptake and metabolism of various carbohydrates30,31. Using the online Motif Alignment & Search Tool (MAST)32, we did not detect MalR1 binding sites, but did identify an possible AraQ binding motif in the promoter region upstream of the serpin-encoding operon (i.e., upstream of BL0109). The resemblance of the AraQ cis-element and the sequence found in the serpin promoter region is modest, but in our opinion warrants further experimental investigation to investigate the proposed involvement of AraQ in serpin regulation in B. longum NCC2705 and other members of the same (sub-)species.
The galactose containing di- and oligosaccharides (i.e. lactose, melibiose and galacto-oligosaccharides [GOS]) as well as the fructose containing di- and oligosaccharides (i.e. saccharose and fructo-oligosaccharides [FOS]) induce serpin production. Importantly, the presence of glucose in the oligosaccharides prevented galactose induction unless the glucose moiety was completely consumed. GOS are produced using enzymatic synthesis from lactose33,34 and usually contain high levels of free lactose and glucose that we demonstrated to suppress serpin production. Conversely, FOS are commonly produced by hydrolysis of long chain inulin and therefore have a very low glucose content34. Intriguingly, glucose repression was not observed when cells were grown on saccharose, although this disaccharide is composed of a glucose and a fructose moiety. In contrast to lactose, hydrolysis of sucrose by sucrose phosphorylase (BL0536; EC 2.4.1.7) results in the formation of fructose and glucose-1-phosphate in the cell. Intriguingly, specific regulatory roles have been suggested for intracellular glucose-1-phosphate, including its proposed role in the regulation of virulence genes in Listeria monocytogenes35.
Lacto-N-tetraose (LNT) contains two galactose, one glucose and one N-Acetyl-galactosamine residues2, and hence was tested for its capacity to induce serpin. At an initial concentration of 0.5 % in the culture medium, LNT supported growth and enabled production of serpin in all tested B. longum subsp. longum strains, indicating a high conservation of the induction mechanism. LNT is likely imported as a tetra saccharide, as the B. longum subsp. longum strains lacks the gene encoding an extracellular lacto-N-biosidase that would be required for this reaction36. Assuming that LNT is imported as a tetra saccharide into the cell, the enzymes involved in its intracellular hydrolysis remain to be identified. However, removal of the terminal glucose moiety may be catalyzed by β-galactosidase. The proposed glucose-mediated catabolite repression may thereby also be transiently activated during LNT metabolization but may have remained unobserved in our experiments due to the low concentration of LNT (0.5%) used. The remaining hydrolysis product (Galβ1–3GlcNAcβ1–3Galβ1) is likely cleaved by the lacto-N-biose phosphorylase (encoded by the gene BL1641) into 2 molecules of galactose and one N-acetyl-galactosamine (39). Although we deciphered the role of glucose and galactose in serpin regulation, a possible involvement of N-acetyl-galactosamine remains to be clarified.
Today, even if FOS and GOS are not the oligosaccharides present in human milk, they are commonly used in infant formula as an affordable alternative to HMOs (including lacto-N-tetraose (LNT)37) with the purpose to drive Bifidobacterium enrichment in the infant gut3–5. Our work suggests that these oligosaccharides could have an impact that goes beyond bifidobacterial growth stimulation, by modifying the in situ expression of a specific molecule (i.e., serpin), which has been proposed to play a role in colonization and immunomodulatory properties of B .longum subsp. longum strains.
Methods
Strains and growth conditions
All strains used in this study (Table S2) were obtained from the Nestle Culture Collection (NCC) (Nestlé Research, Lausanne, Switzerland). Growth was routinely performed using de Man, de Rogosa, Sharpe medium (BD Difco, Franklin Lakes, USA) supplemented with 0.05 % of cysteine (MRSc), at 37 °C in anaerobiosis without agitation. Supplementation of the media with 200 µg/ml of spectinomycin was applied for the serpin overexpressing recombinant B. longum NCC 2705 that harbors pMDY2516. Where relevant, protease addition rates in the medium were the same than previously described13, namely 0.5 mg/ml papain, 0.1 mg/ml trypsin, 0.16 mg/ml chymotrypsin (Sigma-Aldrich Chemie GmbH, Buchs, Switzerland) and 1 mg/ml porcine pancreatic elastase (PPE; MP Biomedicals SARL, Illkirch-Graffenstaden, France). Carbohydrate stock solutions were all prepared at 100 g/L in water and filtered sterilized (glucose, arabinose, ribose, xylose, galactose, fructose, lactose, melibiose, saccharose; Sigma-Aldrich Chemie GmbH), galacto-oligosaccharide (GOS) (King GOS [King-Prebiotics, Yunfu City, China]; Vivinal GOS [FrieslandCampina DOMO Amersfoort, The Netherlands]); bovine milk oligosaccharides BMOS [Société des Produits Nestlé, Vevey, Switzerland4], fructo-oligosaccharides (FOS) (NutraFlora FOS [Ingredion Korea Inc, Gyunggi-do, Korea]; Orafti P95 [BENEO GmbH, Mannheim, Germany]) and human milk oligosaccharides (HMOs) (2'-O-Fucosyllactose (2'FL), Lacto-N-tetraose (LNT), Lacto-N-neotetraose (LNnT), 3'-O-Sialyllactose (3'SL), Difucoxyllactose (DiFL) [Glycom A/S, Lyngby, Denmark]). These carbohydrates were added to a MRSc-based medium containing no carbohydrates (10 g/L bacto proteose peptone, 3, 5 g/L bacto yeast extract, 1 g/L Tween 80 [Chemie Brunschwig, Basel, Switzerland]; 2 g/L di-ammonium hydrogen citrate, 5 g/L sodium acetate, 0.1 g/L magnesium sulphate, 0.05 g/L manganese sulfate, 2 g/L di-sodium phosphate, 0.5 g/L cysteine [Sigma-Aldrich Chemie GmbH]). Two (2) % rate inoculum from an MRSc overnight culture was applied for all growth experiments, which were performed in a BioLector microbioreactor system (m2p-labs GmbH, Baesweiler, Germany), using 48 flowerplate inserted in an anaerobic chamber for 16 to 48h (2ml volume per well, agitation at 600 rpm, CO2 atmosphere, 37 °C). Growth was followed over time by continuous measurement of the scattered light at 620 nm and levels of residual glucose in the culture were determined using the MQuant kit (Sigma-Aldrich Chemie GmbH) according to the manufacturer’s protocol.
Subcellular fractions preparation, crude protein extraction and total protein quantification
To determine the subcellular localization of the serpin, cells were grown to stationary phase (16h) and a cell amount corresponding to 12 units of OD600 was harvested by centrifugation (3500 g, 2 min, 4 °C). The supernatant was filtered through a 0.2 µm pore size filter (Sigma-Aldrich Chemie GmbH), and the filtrate was precipitated with an equal volume of ice-cold acetone and placed on ice for 20 minutes prior to centrifugation (12,000 g, 10 min, 4 °C). The obtained pellet was dissolved with 960 µl of 50mM NaOH (i.e., supernatant fraction). The bacterial pellet was washed once with one volume of ice-cold PBS and resuspended in 960 µl of PBS containing 0.1 mg/ml DNAse I, 20 μg/ml Rnase A and SigmaFast Protease Inhibitor (Sigma-Aldrich Chemie GmbH; 1 tablet per 100 ml of solution). The obtained solution was incubated at 37 °C for 30 minutes and was subsequently lysed by bead-beating using a FastPrep-24 (MP Biomedicals SARL), 3 times for 1 minute at 4 m/s, with 2 minutes cooling on ice in between. The obtained crude cell lysate was separated in cytoplasmic (soluble) and crude cell wall (pellet) fractions by centrifugation (12,000 g, 10 min, 4 °C). The cell wall fraction pellet was further rinsed twice and resuspended in 960 µl of PBS containing proteinase inhibitor, which was tested to not interfere with the developed ELISA. All fractions were analyzed for serpin content (see below) and obtained values were normalized based on culture optical density, measured at 600 nm (OD600).
For all other analyses, stationary phase bacterial cultures (after 16h of growth, unless stated otherwise) were harvested by centrifugation (3500 g, 2 min, 4 °C). In order to quantify residual glucose content, supernatants were filter sterilized using a 0.45 µm filter and stored at − 20 °C. Bacterial pellets were washed with one volume of Dulbecco’s Phosphate Buffer (PBS; Sigma-Aldrich Chemie GmbH) and resuspended in 600 µl of PBS containing Halt Protease Inhibitor (Sigma-Aldrich Chemie GmbH). Bacteria were subsequently lysed by bead-beating using a FastPrep-24 (see above; MP Biomedicals SARL). Crude lysates were used as such, containing both soluble and non-soluble fractions. Total protein content was determined using the Pierce BCA protein Assay kit (Thermo Fisher Scientific AG, Basel, Switzerland).
Serpin protein quantification by sandwich ELISA
All anti-serpin rabbit polyclonal antibodies used in this work were obtained from Proteogenix (Schiltigheim, France) and using a purified B. longum NCC 2705 recombinant serpin produced in Escherichia coli12. For the sandwich ELISA, 96 well plates (Nunc MaxiSorp [Thermo Fisher Scientific AG]) were used. Coating was performed for 16h at 4 °C using 100 µl per well of a 0.2 M sodium carbonate/biocarbonate solution at pH 9.4 containing 250 ng/ml of primary anti-serpin antibodies. In between each step, plates were washed 3 times using 300 µl of wash buffer (WB; PBS to which 0.05% of Tween 20 was added). Blocking was performed for 1h at room temperature using 300 µl of blocking buffer (BB; WB to which 2% of Bovine Serum Albumin [BSA; Sigma-Aldrich Chemie GmbH] was added). The standard (purified recombinant serpin12) and the different samples were diluted in a final volume of 100 µl of BB and incubated for 2h at room temperature. Secondary biotinylated anti-serpin antibody (250 ng/ml in 100 µl of WB containing 0.2% BSA) was added and incubated for 1h at room temperature, followed by the addition of 100 µl of the enzyme conjugate (HRP Pierce, Thermo Fisher Scientific AG) (10 µg/ml in WB containing 0.2% BSA) and continuing incubation for 1h at room temperature. Subsequently, repeated (6) washes using 300 µl of WB were performed to remove background reactivity. Detection was performed by adding 100 µl per well of HRP substrate (1-step Ultra TMB ELISA, ThermoFisher Scientific AG) and 15 minutes incubation at room temperature. The reaction was terminated by the addition of 100 µl of 2M sulfuric acid and final optical density at 450 nm was measured in a Varioskan spectrophotometer (Thermo Fisher Scientific AG).
Standard curves were obtained following the sigmoidal 4 parameter logistic regression (4PL) method and concentrations of serpin in individual samples were calculated using the following formula:
A: Minimal asymptote (); B: Slope; C: Inflection point (); D: Maximal asymptote ().
Statistical analysis
All data obtained for B. longum NCC 2705 grown in either MRSc or MRSc + 1% glucose were pooled—(n=36) and were analyzed for normality using D’Agostino and Pearson algorithm (data not shown). This data pool was used as control group in every analysis. Data were log transformed to compensate for potential heteroscedasticity and one-way ANOVA was performed on every dataset. P-values were obtained by performing a Turkey’s multiple comparison with an alpha level of 0.05.
Bioinformatic analysis
All gene and protein sequences of B. longum NCC 2705 referred to in this work were obtained from the National Center for Biotechnology Information (NCBI; NC_004307.2). Operon prediction was obtained from the Prokaryotic Operon Database (ProOpDB)38. Functional annotation of the B. longum NCC 2705 serpin protein encoded by the BL0108 gene was performed using Interpro-scan20 and signal peptide prediction was performed using Signal P21.
To shed light on the genetic mechanism involved in regulation of serpin production, all bindings sites of transcriptional factors known to be implicated in carbohydrate metabolism regulation were retrieved from the RegPrecise database29. These sequences were used as input in the “Motif Alignment and Search Tool” (MAST)32, to identify resembling motifs in the region upstream of the Bl0109 gene (genome positions 132852 to 134216; reverse sequenced). Obtained candidate motif-homologues were subsequently mapped relative to the serpin promoter elements (i.e., the -35 and -10 regions) that were previously described by Turroni et al.13.
Supplementary Information
Acknowledgements
We thank Bastien Gentili for his contribution to experimental work and Biljana Bogicevic for her inputs in the final revision of this manuscript.
Author contributions
S.D. conceived, planned and carried out all experiments under supervision of M.K., M.G. developed the serpin-specific ELISA assay. S.D., A.M., and M.K. took the lead in writing the manuscript. All authors, including J.A.M., G.B., and C.J.B. provided critical feedback and helped shape the research, analysis of results and manuscript.
Competing interests
S. Duboux, M. Golliard, J.A. Muller, G. Bergonzelli and C.J Bolten are employed by Nestlé (Société des Produits Nestlé SA).
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
S. Duboux, Email: stephane.duboux@rdls.nestle.com
M. Kleerebezem, Email: michiel.kleerebezem@wur.nl
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-86740-y.
References
- 1.Turroni F, et al. Bifidobacteria and the infant gut: an example of co-evolution and natural selection. Cell Mol. Life Sci. 2018;75:103–118. doi: 10.1007/s00018-017-2672-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayechu-Muruzabal V, et al. Diversity of human milk oligosaccharides and effects on early life immune development. Front. Pediatr. 2018;6:239. doi: 10.3389/fped.2018.00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jakobsen LMA, Sundekilde UK, Andersen HJ, Nielsen DS, Bertram HC. Lactose and bovine milk oligosaccharides synergistically stimulate b. longum subsp. longum growth in a simplified model of the infant gut microbiome. J. Proteome Res. 2019 doi: 10.1021/acs.jproteome.9b00211. [DOI] [PubMed] [Google Scholar]
- 4.Meli F, et al. Growth and safety evaluation of infant formulae containing oligosaccharides derived from bovine milk: a randomized, double-blind, noninferiority trial. BMC Pediatr. 2014;14:306. doi: 10.1186/s12887-014-0306-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tandon D, et al. A prospective randomized, double-blind, placebo-controlled, dose-response relationship study to investigate efficacy of fructo-oligosaccharides (FOS) on human gut microflora. Sci. Rep. 2019;9:5473. doi: 10.1038/s41598-019-41837-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hidalgo-Cantabrana C, et al. Bifidobacteria and their health-promoting effects. Microbiol. Spectr. 2017 doi: 10.1128/microbiolspec.BAD-0010-2016. [DOI] [PubMed] [Google Scholar]
- 7.Schell MA, et al. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. USA. 2002;99:14422–14427. doi: 10.1073/pnas.212527599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parche S, et al. Sugar transport systems of Bifidobacterium longum NCC2705. J. Mol. Microbiol. Biotechnol. 2007;12:9–19. doi: 10.1159/000096455. [DOI] [PubMed] [Google Scholar]
- 9.Gueimonde M, Garrigues C, van Sinderen D, de los Reyes-Gavilan CG, Margolles A. Bile-inducible efflux transporter from Bifidobacterium longum NCC2705 conferring bile resistance. Appl. Environ. Microbiol. 2009;75:3153–3160. doi: 10.1128/AEM.00172-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki K, et al. Adhesion properties of a putative polymorphic fimbrial subunit protein from Bifidobacterium longum subsp. longum. Biosci. Microbiota Food Health. 2016;35:19–27. doi: 10.12938/bmfh.2015-015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukuda S, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 12.Ivanov D, et al. A serpin from the gut bacterium Bifidobacterium longum inhibits eukaryotic elastase-like serine proteases. J. Biol. Chem. 2006;281:17246–17252. doi: 10.1074/jbc.M601678200. [DOI] [PubMed] [Google Scholar]
- 13.Turroni F, et al. Characterization of the serpin-encoding gene of Bifidobacterium breve 210B. Appl. Environ. Microbiol. 2010;76:3206–3219. doi: 10.1128/AEM.02938-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riedel CU, et al. Anti-inflammatory effects of bifidobacteria by inhibition of LPS-induced NF-kappaB activation. World J. Gastroenterol. 2006;12:3729–3735. doi: 10.3748/wjg.v12.i23.3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burg ND, Pillinger MH. The neutrophil: function and regulation in innate and humoral immunity. Clin. Immunol. 2001;99:7–17. doi: 10.1006/clim.2001.5007. [DOI] [PubMed] [Google Scholar]
- 16.McCarville JL, et al. A commensal Bifidobacterium longum strain improves gluten-related immunopathology in mice through expression of a serine protease inhibitor. Appl. Environ. Microbiol. 2017 doi: 10.1128/AEM.01323-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buhner S, et al. Protease signaling through protease activated receptor 1 mediate nerve activation by mucosal supernatants from irritable bowel syndrome but not from ulcerative colitis patients. PLoS ONE. 2018;13:e0193943. doi: 10.1371/journal.pone.0193943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Leary NA, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucl. Acids Res. 2016;44:D733–745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alvarez-Martin P, et al. A two-component regulatory system controls autoregulated serpin expression in Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 2012;78:7032–7041. doi: 10.1128/AEM.01776-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitchell AL, et al. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucl. Acids Res. 2019;47:D351–D360. doi: 10.1093/nar/gky1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Almagro Armenteros JJ, et al. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019;37:420–423. doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- 22.Pokusaeva K, Fitzgerald GF, van Sinderen D. Carbohydrate metabolism in Bifidobacteria. Genes Nutr. 2011;6:285–306. doi: 10.1007/s12263-010-0206-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parche S, et al. Lactose-over-glucose preference in Bifidobacterium longum NCC2705: glcP, encoding a glucose transporter, is subject to lactose repression. J. Bacteriol. 2006;188:1260–1265. doi: 10.1128/JB.188.4.1260-1265.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei X, et al. Fructose uptake in Bifidobacterium longum NCC2705 is mediated by an ATP-binding cassette transporter. J. Biol. Chem. 2012;287:357–367. doi: 10.1074/jbc.M111.266213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deutscher J. The mechanisms of carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 2008;11:87–93. doi: 10.1016/j.mib.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 26.Caescu CI, Vidal O, Krzewinski F, Artenie V, Bouquelet S. Bifidobacterium longum requires a fructokinase (Frk; ATP:D-fructose 6-phosphotransferase, EC 2.7.1.4) for fructose catabolism. J. Bacteriol. 2004;186:6515–6525. doi: 10.1128/JB.186.19.6515-6525.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trindade MI, Abratt VR, Reid SJ. Induction of sucrose utilization genes from Bifidobacterium lactis by sucrose and raffinose. Appl. Environ. Microbiol. 2003;69:24–32. doi: 10.1128/aem.69.1.24-32.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nessler S, et al. HPr kinase/phosphorylase, the sensor enzyme of catabolite repression in Gram-positive bacteria: structural aspects of the enzyme and the complex with its protein substrate. J. Bacteriol. 2003;185:4003–4010. doi: 10.1128/jb.185.14.4003-4010.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Novichkov PS, et al. RegPrecise 3.0: a resource for genome-scale exploration of transcriptional regulation in bacteria. BMC Genomics. 2013;14:745. doi: 10.1186/1471-2164-14-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lanigan N, et al. Transcriptional control of central carbon metabolic flux in Bifidobacteria by two functionally similar, yet distinct LacI-type regulators. Sci. Rep. 2019;9:17851. doi: 10.1038/s41598-019-54229-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khoroshkin MS, Leyn SA, Van Sinderen D, Rodionov DA. Transcriptional regulation of carbohydrate utilization pathways in the bifidobacterium genus. Front. Microbiol. 2016;7:120. doi: 10.3389/fmicb.2016.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bailey TL, et al. MEME SUITE: tools for motif discovery and searching. Nucl. Acids Res. 2009;37:W202–208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ganzle M. Enzymatic synthesis of galacto-oligosaccharides and other lactose derivatives (hetero-oligosaccharides) from lactose. Int. Dairy J. 2001;22:116–122. doi: 10.1016/j.idairyj.2011.06.010. [DOI] [Google Scholar]
- 34.Martins GN, Ureta MM, Tymczyszyn EE, Castilho PC, Gomez-Zavaglia A. Technological aspects of the production of fructo and galacto-oligosaccharides: enzymatic synthesis and hydrolysis. Front. Nutr. 2019;6:78. doi: 10.3389/fnut.2019.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ripio MT, Brehm K, Lara M, Suarez M, Vazquez-Boland JA. Glucose-1-phosphate utilization by Listeria monocytogenes is PrfA dependent and coordinately expressed with virulence factors. J. Bacteriol. 1997;179:7174–7180. doi: 10.1128/jb.179.22.7174-7180.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakurama H, et al. Lacto-N-biosidase encoded by a novel gene of Bifidobacterium longum subspecies longum shows unique substrate specificity and requires a designated chaperone for its active expression. J. Biol. Chem. 2013;288:25194–25206. doi: 10.1074/jbc.M113.484733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kunz C, Rudloff S, Baier W, Klein N, Strobel S. Oligosaccharides in human milk: structural, functional, and metabolic aspects. Annu. Rev. Nutr. 2000;20:699–722. doi: 10.1146/annurev.nutr.20.1.699. [DOI] [PubMed] [Google Scholar]
- 38.Taboada B, Ciria R, Martinez-Guerrero CE, Merino E. ProOpDB: prokaryotic operon database. Nucl. Acids Res. 2012;40:D627–631. doi: 10.1093/nar/gkr1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.