Fig. 1. Residues of 114–130 of the RelA pseudo-hydrolase domain form a loop that controls ppGpp synthesis.
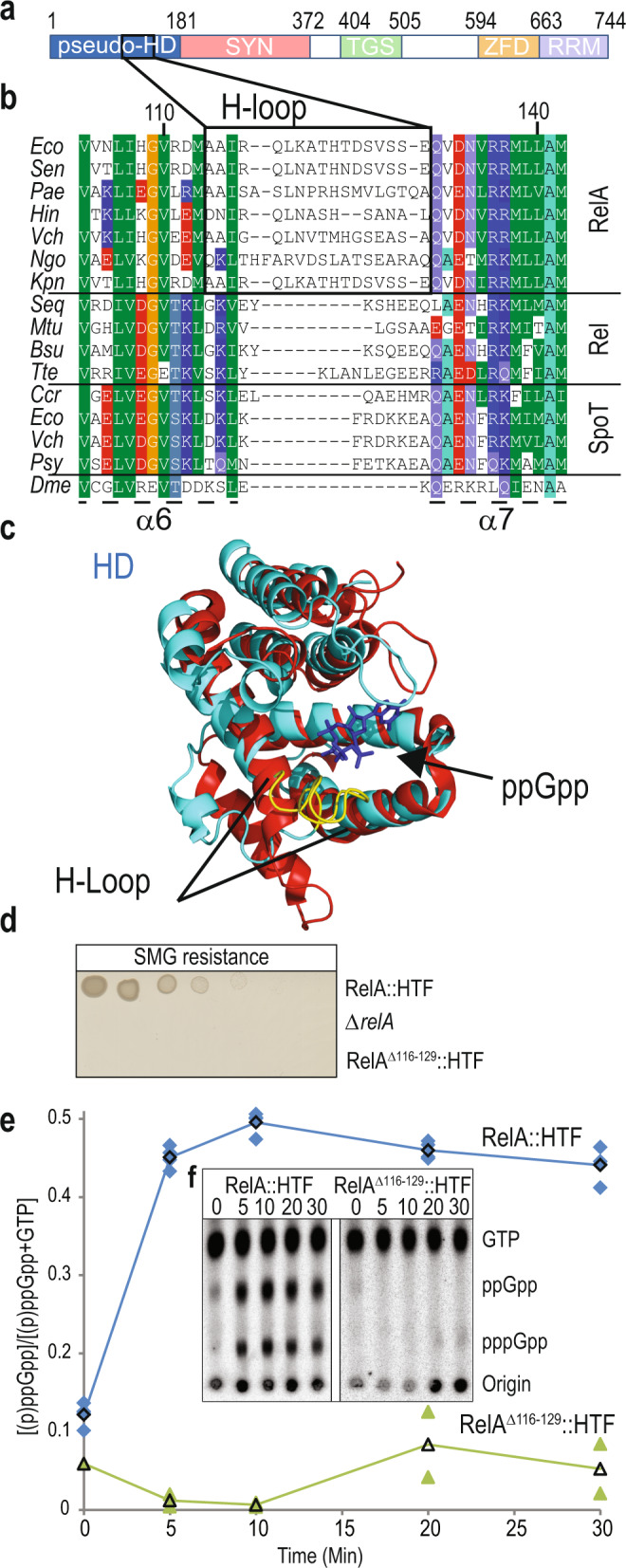
a Illustration of functional domains of RelA: HYD inactive hydrolase domain (residues 1–181), SYN synthetase domain (residues 182–372), TGS ThrRS, GTPase and SpoT-like domain (residues 404–505), ZFD Zinc-finger domain (residues 594–663) and RRM RNA recognition motif (residues 664–744). b Multiple sequence alignment of selected RelA, Rel and SpoT sequences. Eco Escherichia coli (RelA: NP_417264.1, SpoT: NP_418107.1), Sen Salmonella enterica (NP_461877.1), Pae Pseudomonas aeruginosa (NP_249625.1), Hin Hemophilus influenza (WP_011271992.1), Vch Vibrio cholera (RelA: WP_000226858.1, SpoT: WP_010895463.1), Ngo Neisseria gonorrhoeae (AP023075.1), Kpn Klebsiella pneumoniae (CP006918.1), Seq Streptococcus dysgalactiae subsp. equisimilis (Q54089), Mtu Mycobacterium tuberculosis (NP_217099), Bsu Bacillus subtilis (NP_390638),Tte Thermus thermophilus (WP_011173739.1), Ccr Caulobacter crescentus (WP_010919427.1), Psy Pseudomonas syringae (WP_003096603.1), Dme Drosophila melanogaster (NP_651682.1). Location of helices α6, α7 and the H-loop is indicated. c Structure of RelA pseudo-hydrolase domain (PDB: 5IQR, shown in cyan) superimposed onto RelTte hydrolase domain (HD, PDB: 6S2T, shown in red). Position of ppGpp in RelTte hydrolase active site is indicated in blue and the H-loop of RelA is shown in yellow. d Functional assay of stringent response in H-loop deletion mutant RelAΔ116–129. Escherichia coli K-12 MG1655 relA::HTF, MG1655 ΔrelA and MG1655 relAΔ116–129::HTF were grown overnight in LB medium at 37 °C. The cells were washed and serial diluted in phosphate buffered saline (PBS) and spotted onto MOPS MM SMG agar plates (SMG resistance) and plates were incubated at 30 °C (See Supplementary Fig. 1f for loading controls). e, f (p)ppGpp measurements in RelA H-loop mutant. Strains from d were grown exponentially in MOPS minimal medium at 30 °C containing [32P]-radiolabeled phosphate as described in methods. At time zero cells were starved for isoleucine by addition of 500 μg/mL L-Valine (final concentration). Samples were collected at time points indicated (min), precipitated in formic acid and spotted on a TLC plate. Nucleotides were separated using 1.5 M potassium phosphate pH 3.4 as solvent. e Quantification of (p)ppGpp for RelA::HTF (n = 4 biologically independent samples are shown with blue diamonds,) and RelAΔ116–129::HTF (n = 2 biologically independent samples are shown with Green triangles). The curves with black symbols indicate averaged values. f Representative TLC from e), positions of GTP, ppGpp and pppGpp are indicated. For TLCs of biological replicates see Supplementary Fig. 1i–n.
